2EVE
 
 | | X-Ray Crystal Structure of Protein PSPTO5229 from Pseudomonas syringae. Northeast Structural Genomics Consortium Target PsR62 | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, ... | | Authors: | Forouhar, F, Zhou, W, Belachew, A, Jayaraman, S, Ciao, M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-08 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural genomics reveals EVE as a new ASCH/PUA-related domain.
Proteins, 75, 2009
|
|
2FFM
 
 | | X-Ray Crystal Structure of Protein SAV1430 from Staphylococcus aureus. Northeast Structural Genomics Consortium Target ZR18. | | Descriptor: | SAV1430 | | Authors: | Forouhar, F, Chen, Y, Jayaraman, S, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-19 | | Release date: | 2005-12-27 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of the Hypothetical Protein SAV1430 from Staphylococcus aureus, Northeast Structural Genomics ZR18.
To be Published
|
|
2P6Y
 
 | | X-ray structure of the protein Q9KM02_VIBCH from Vibrio cholerae at the resolution 1.63 A. Northeast Structural Genomics Consortium target VcR80. | | Descriptor: | Hypothetical protein VCA0587, ZINC ION | | Authors: | Kuzin, A.P, Abashidze, M, Jayaraman, S, Chen, C.X, Wang, C, Fang, Y, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-03-19 | | Release date: | 2007-06-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray structure of the protein Q9KM02_VIBCH from Vibrio cholerae at the resolution 1.63 A.
To be Published
|
|
3JCZ
 
 | | Structure of bovine glutamate dehydrogenase in the unliganded state | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-27 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3J7H
 
 | | Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy | | Descriptor: | Beta-galactosidase, MAGNESIUM ION | | Authors: | Bartesaghi, A, Matthies, D, Banerjee, S, Merk, A, Subramaniam, S. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of beta-galactosidase at 3.2- angstrom resolution obtained by cryo-electron microscopy.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3JD2
 
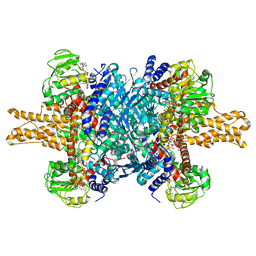 | | Glutamate dehydrogenase in complex with NADH, open conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD0
 
 | | Glutamate dehydrogenase in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD1
 
 | | Glutamate dehydrogenase in complex with NADH, closed conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
2GSW
 
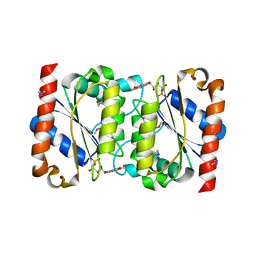 | | Crystal Structure of the Putative NADPH-dependent Azobenzene FMN-Reductase YhdA from Bacillus subtilis, Northeast Structural Genomics Target SR135 | | Descriptor: | FLAVIN MONONUCLEOTIDE, yhdA | | Authors: | Forouhar, F, Hussain, M, Jayaraman, S, Shen, J, Cooper, B, Cunningham, K, Janjua, H, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Crystal Structure of the Putative NADPH-dependent Azobenzene FMN-Reductase YhdA from Bacillus subtilis, Northeast Structural Genomics Target SR135
To be Published
|
|
2GO8
 
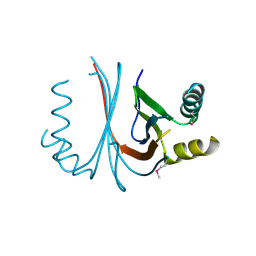 | | Crystal structure of YQJZ_BACSU FROM Bacillus subtilis. Northeast structural genomics TARGET SR435 | | Descriptor: | Hypothetical protein yqjZ | | Authors: | Benach, J, Su, M, Jayaraman, S, Fang, Y, Xiao, R, Ma, L.-C, Cunningham, K, Wang, D, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-12 | | Release date: | 2006-04-25 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of YQJZ_BACSU from Bacillus subtilis. Northeast Structural Genomics TARGET SR435
To be Published
|
|
4IFF
 
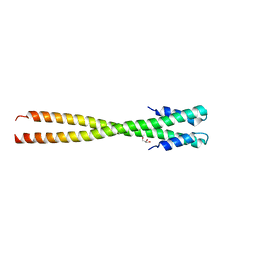 | | Structural organization of FtsB, a transmembrane protein of the bacterial divisome | | Descriptor: | Fusion of phage phi29 Gp7 protein and Cell division protein FtsB, GLYCEROL | | Authors: | LaPointe, L.M, Taylor, K.C, Subramaniam, S, Khadria, A, Rayment, I, Senes, A. | | Deposit date: | 2012-12-14 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Organization of FtsB, a Transmembrane Protein of the Bacterial Divisome.
Biochemistry, 52, 2013
|
|
3JCF
 
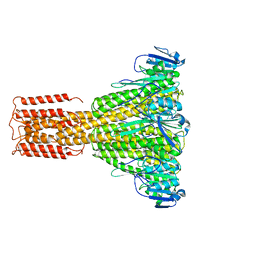 | |
3JCG
 
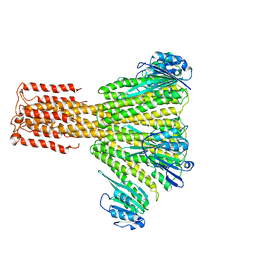 | |
3JCH
 
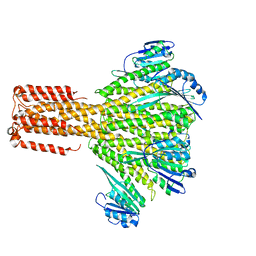 | |
2QGM
 
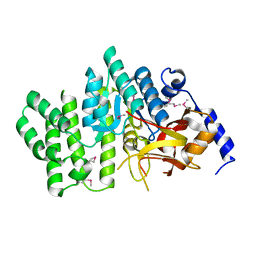 | | Crystal structure of succinoglycan biosynthesis protein at the resolution 1.7 A. Northeast Structural Genomics Consortium target BcR136. | | Descriptor: | Succinoglycan biosynthesis protein | | Authors: | Kuzin, A.P, Abashidze, M, Jayaraman, S, Wang, H, Fang, Y, Maglaqui, M, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-24 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of succinoglycan biosynthesis protein at the resolution 1.7 A. Northeast Structural Genomics Consortium target BcR136.
To be Published
|
|
2QGG
 
 | | X-Ray structure of the protein Q6F7I0 from Acinetobacter calcoaceticus AmMS 248. Northeast Structural Genomics Consortium target AsR73. | | Descriptor: | 16S rRNA-processing protein rimM, POTASSIUM ION, UNKNOWN LIGAND | | Authors: | Kuzin, A.P, Su, M, Jayaraman, S, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-Ray structure of the protein Q6F7I0 from Acinetobacter calcoaceticus AmMS 248.
To be Published
|
|
2QGU
 
 | | Three-dimensional structure of the phospholipid-binding protein from Ralstonia solanacearum Q8XV73_RALSQ in complex with a phospholipid at the resolution 1.53 A. Northeast Structural Genomics Consortium target RsR89 | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Probable signal peptide protein | | Authors: | Kuzin, A.P, Chen, Y, Jayaraman, S, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional structure of the phospholipid-binding protein from Ralstonia solanacearum Q8XV73_RALSQ in complex with a phospholipid at the resolution 1.53 A.
To be Published
|
|
2IEE
 
 | | Crystal Structure of YCKB_BACSU from Bacillus subtilis. Northeast Structural Genomics Consortium target SR574. | | Descriptor: | Probable ABC transporter extracellular-binding protein yckB | | Authors: | Kuzin, A.P, Su, M, Jayaraman, S, Chen, X.C, Jang, M, Cunningham, K, Ma, C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-18 | | Release date: | 2006-10-03 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of probable ABC transporter extracellular-binding protein yckB from Bacillus subtilis.
To be Published
|
|
3EAJ
 
 | |
3E91
 
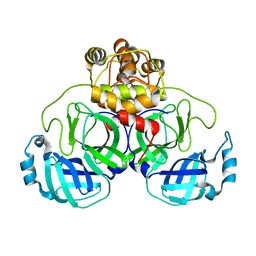 | |
3EA9
 
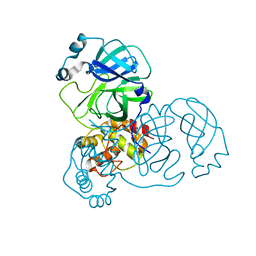 | |
3EA8
 
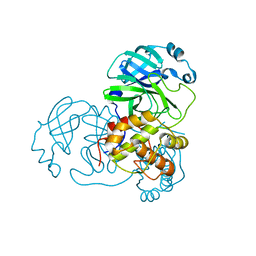 | |
3EA7
 
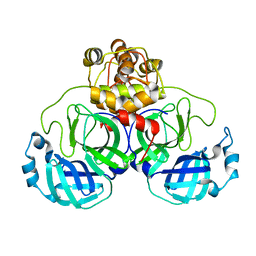 | |
3DNO
 
 | | Molecular structure for the HIV-1 gp120 trimer in the CD4-bound state | | Descriptor: | HIV-1 envelope glycoprotein gp120 | | Authors: | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | Deposit date: | 2008-07-02 | | Release date: | 2008-08-19 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
3DNN
 
 | | Molecular structure for the HIV-1 gp120 trimer in the unliganded state | | Descriptor: | HIV-1 envelope glycoprotein gp120 | | Authors: | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | Deposit date: | 2008-07-02 | | Release date: | 2008-08-19 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
