3RP1
 
 | | Crystal structure of Human LAIR-1 in C2 space group | | Descriptor: | Leukocyte-associated immunoglobulin-like receptor 1 | | Authors: | Sampathkumar, P, Ramagopal, U.A, Yan, Q, Toro, R, Nathenson, S, Bonanno, J, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-26 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Human LAIR-1 in C2 space group
To be Published
|
|
3OF3
 
 | | Crystal structure of PNP with an inhibitor DADME_immH from Vibrio cholerae | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type 1 | | Authors: | Kim, J, Ramagopal, U.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-13 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of PNP with an inhibitor DADME_immH from Vibrio cholerae
To be Published
|
|
3N07
 
 | | Structure of putative 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Vibrio cholerae | | Descriptor: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, MAGNESIUM ION | | Authors: | Liu, W, Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-13 | | Release date: | 2010-08-04 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
3OHP
 
 | |
3RRA
 
 | | Crystal structure of enolase PRK14017 (target EFI-500653) from Ralstonia pickettii 12J with magnesium bound | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative D-galactonate dehydratase | | Authors: | Patskovsky, Y, Ramagopal, U.A, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of enolase PRK14017 from Ralstonia pickettii
To be Published
|
|
3RRQ
 
 | |
3MHD
 
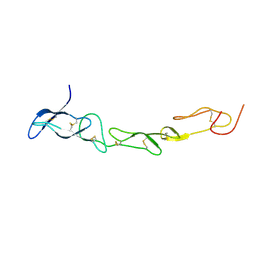 | | Crystal structure of DCR3 | | Descriptor: | Tumor necrosis factor receptor superfamily member 6B | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-04-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
3MI8
 
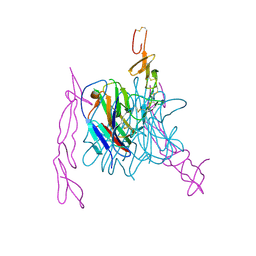 | | The structure of TL1A-DCR3 COMPLEX | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 15, SECRETED FORM, Tumor necrosis factor receptor superfamily member 6B | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-04-09 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
3RBG
 
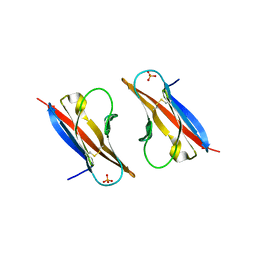 | | Crystal structure analysis of Class-I MHC restricted T-cell associated molecule | | Descriptor: | Cytotoxic and regulatory T-cell molecule, PHOSPHATE ION | | Authors: | Rubinstein, R, Ramagopal, U.A, Toro, R, Nathenson, S.G, Fiser, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2011-03-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional classification of immune regulatory proteins.
Structure, 21, 2013
|
|
3RNQ
 
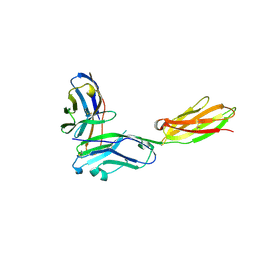 | |
3RNK
 
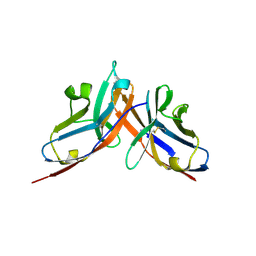 | | Crystal structure of the complex between mouse PD-1 mutant and PD-L2 IgV domain | | Descriptor: | Programmed cell death 1 ligand 2, Programmed cell death protein 1 | | Authors: | Lazar-Molnar, E, Ramagopal, U.A, Cao, E, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2011-04-22 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of the complex between mouse PD-1 mutant and PD-L2 IgV domain
To be Published
|
|
3OCC
 
 | | Crystal structure of PNP with DADMEimmH from Yersinia pseudotuberculosis | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | Authors: | Kim, J, Ramagopal, U.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-09 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | crystal structure of PNP with DADMEimmH from Yersinia pseudotuberculosis
To be Published
|
|
3SBW
 
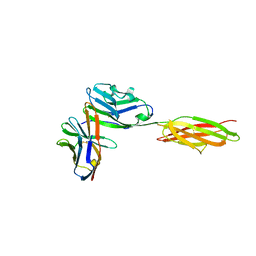 | | Crystal structure of the complex between the extracellular domains of mouse PD-1 mutant and human PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | Authors: | Lazar-Molnar, E, Ramagopal, U.A, Cao, E, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2011-06-06 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the complex between the extracellular domains of mouse PD-1 mutant and human PD-L1
To be published
|
|
3GMF
 
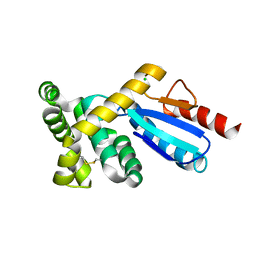 | | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans | | Descriptor: | CHLORIDE ION, Protein-disulfide isomerase | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans
To be Published
|
|
3G7U
 
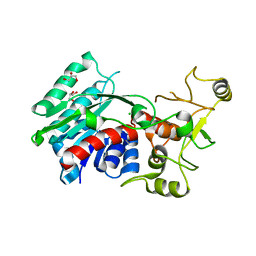 | | Crystal structure of putative DNA modification methyltransferase encoded within prophage Cp-933R (E.coli) | | Descriptor: | CHLORIDE ION, Cytosine-specific methyltransferase, GLYCEROL | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Gilmore, M, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-10 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of DNA Modification Methyltransferase Encoded within Prophage Cp-933R (E.coli)
To be Published
|
|
3GD5
 
 | | Crystal structure of ornithine carbamoyltransferase from Gloeobacter violaceus | | Descriptor: | Ornithine carbamoyltransferase | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Ramagopal, U.A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-23 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of ornithine carbamoyltransferase from Gloeobacter violaceus
To be Published
|
|
3GRZ
 
 | | CRYSTAL STRUCTURE OF ribosomal protein L11 methylase FROM Lactobacillus delbrueckii subsp. bulgaricus | | Descriptor: | GLYCEROL, Ribosomal protein L11 methyltransferase | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN 11 METHYLASE FROM Lactobacillus delbrueckii subsp. bulgaricus
To be Published
|
|
3K12
 
 | | Crystal structure of an uncharacterized protein A6V7T0 from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, uncharacterized protein A6V7T0 | | Authors: | Ho, M, Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-27 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of an uncharacterized protein A6V7T0 from Pseudomonas aeruginosa
To be published
|
|
3OCQ
 
 | |
3ODG
 
 | | crystal structure of xanthosine phosphorylase bound with xanthine from Yersinia pseudotuberculosis | | Descriptor: | CHLORIDE ION, XANTHINE, Xanthosine phosphorylase | | Authors: | Kim, J, Ramagopal, U.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-11 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | crystal structure of xanthosine phosphorylase bound with xanthine from Yersinia pseudotuberculosis
To be Published
|
|
3C3M
 
 | | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1 | | Descriptor: | GLYCEROL, Response regulator receiver protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Dickey, M, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1.
To be Published
|
|
3EEY
 
 | | CRYSTAL STRUCTURE OF PUTATIVE RRNA-METHYLASE FROM Clostridium thermocellum | | Descriptor: | GLYCEROL, Putative rRNA methylase, S-ADENOSYLMETHIONINE, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Rutter, M, Hu, S, Bain, K, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-06 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Rrna-Methylase from Clostridium Thermocellum
To be Published
|
|
3CJP
 
 | | Crystal structure of an uncharacterized amidohydrolase CAC3332 from Clostridium acetobutylicum | | Descriptor: | Predicted amidohydrolase, dihydroorotase family, ZINC ION | | Authors: | Malashkevich, V.N, Toro, R, Ramagopal, U.A, Bonanno, J.B, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-13 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of an uncharacterized amidohydrolase CAC3332 from Clostridium acetobutylicum.
To be Published
|
|
3CFY
 
 | | Crystal structure of signal receiver domain of putative Luxo repressor protein from Vibrio parahaemolyticus | | Descriptor: | Putative LuxO repressor protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Fong, R, Freeman, J, Iizuka, M, Groshong, C, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Putative Luxo Repressor Protein from Vibrio Parahaemolyticus.
To be Published
|
|
3E05
 
 | | CRYSTAL STRUCTURE OF Precorrin-6y C5,15-methyltransferase FROM Geobacter metallireducens GS-15 | | Descriptor: | CHLORIDE ION, GLYCEROL, Precorrin-6Y C5,15-methyltransferase (Decarboxylating) | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Dickey, M, Hu, S, Maletic, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-30 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF Precorrin-6y C5,15-methyltransferase from Geobacter metallireducens
To be Published
|
|
