4JQW
 
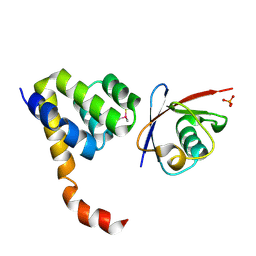 | | Crystal Structure of a Complex of NOD1 CARD and Ubiquitin | | Descriptor: | Nucleotide-binding oligomerization domain-containing protein 1, PHOSPHATE ION, Polyubiquitin-C | | Authors: | Ver Heul, A.M, Gakhar, L, Piper, R.C, Ramaswamy, S. | | Deposit date: | 2013-03-20 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a complex of NOD1 CARD and ubiquitin
Plos One, 9, 2014
|
|
3MCJ
 
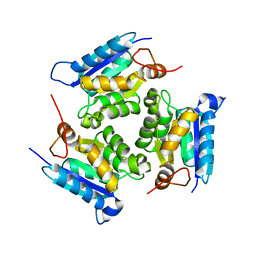 | | Crystal structure of molybdenum cofactor biosynthesis (AQ_061) other form from aquifex aeolicus VF5 | | Descriptor: | 1,2-ETHANEDIOL, Molybdenum cofactor biosynthesis MOG | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-03-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures, dynamics and functional implications of molybdenum-cofactor biosynthesis protein MogA from two thermophilic organisms
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2CB1
 
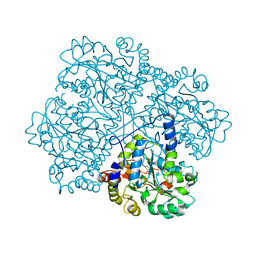 | | Crystal Structure of O-actetyl Homoserine Sulfhydrylase From Thermus Thermophilus HB8,OAH2. | | Descriptor: | O-ACETYL HOMOSERINE SULFHYDRYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Imagawa, T, Utsunomiya, H, Tsuge, H, Ebihara, A, Kanagawa, M, Nakagawa, N, Kuroishi, C, Agari, Y, Kuramitsu, S, Yokoyama, S. | | Deposit date: | 2005-12-28 | | Release date: | 2007-01-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of O-Acetyl Homoserine Sulfhydrylase
To be Published
|
|
4HM2
 
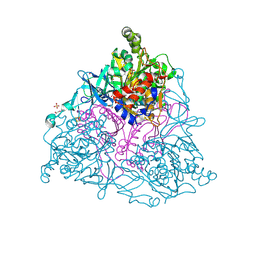 | | Naphthalene 1,2-Dioxygenase bound to ethylphenylsulfide | | Descriptor: | (ethylsulfanyl)benzene, 1,2-ETHANEDIOL, FE (III) ION, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | One enzyme, many reactions: structural basis for the various reactions catalyzed by naphthalene 1,2-dioxygenase.
Iucrj, 4, 2017
|
|
2QGM
 
 | | Crystal structure of succinoglycan biosynthesis protein at the resolution 1.7 A. Northeast Structural Genomics Consortium target BcR136. | | Descriptor: | Succinoglycan biosynthesis protein | | Authors: | Kuzin, A.P, Abashidze, M, Jayaraman, S, Wang, H, Fang, Y, Maglaqui, M, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of succinoglycan biosynthesis protein at the resolution 1.7 A. Northeast Structural Genomics Consortium target BcR136.
To be Published
|
|
4HJL
 
 | | Naphthalene 1,2-Dioxygenase bound to 1-chloronaphthalene | | Descriptor: | 1,2-ETHANEDIOL, 1-chloronaphthalene, FE (III) ION, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-12 | | Release date: | 2013-10-16 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | One enzyme, many reactions: structural basis for the various reactions catalyzed by naphthalene 1,2-dioxygenase.
Iucrj, 4, 2017
|
|
4HM7
 
 | | Naphthalene 1,2-Dioxygenase bound to styrene | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | One enzyme, many reactions: structural basis for the various reactions catalyzed by naphthalene 1,2-dioxygenase.
Iucrj, 4, 2017
|
|
4HM3
 
 | | Naphthalene 1,2-Dioxygenase bound to ethylbenzene | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | One enzyme, many reactions: structural basis for the various reactions catalyzed by naphthalene 1,2-dioxygenase.
Iucrj, 4, 2017
|
|
4HM5
 
 | | Naphthalene 1,2-Dioxygenase bound to indene | | Descriptor: | 1,2-ETHANEDIOL, 2,3-dihydro-1H-indene, FE (III) ION, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | One enzyme, many reactions: structural basis for the various reactions catalyzed by naphthalene 1,2-dioxygenase.
Iucrj, 4, 2017
|
|
2YS7
 
 | | Crystal structure of GAR synthetase from Geobacillus kaustophilus | | Descriptor: | Phosphoribosylglycinamide synthetase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
4HM1
 
 | | Naphthalene 1,2-Dioxygenase bound to 1-indanone | | Descriptor: | 1,2-ETHANEDIOL, 2,3-dihydro-1H-inden-1-one, FE (III) ION, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Naphthalene 1,2-Dioxygenase bound to 1-indanone
To be Published
|
|
4HKV
 
 | | Naphthalene 1,2-Dioxygenase bound to benzamide | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDE, FE (III) ION, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-15 | | Release date: | 2013-10-30 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | One enzyme, many reactions: structural basis for the various reactions catalyzed by naphthalene 1,2-dioxygenase.
Iucrj, 4, 2017
|
|
4HM0
 
 | | Naphthalene 1,2-Dioxygenase bound to indole-3-acetate | | Descriptor: | 1,2-ETHANEDIOL, 1H-INDOL-3-YLACETIC ACID, FE (III) ION, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | One enzyme, many reactions: structural basis for the various reactions catalyzed by naphthalene 1,2-dioxygenase.
Iucrj, 4, 2017
|
|
4HM4
 
 | | Naphthalene 1,2-Dioxygenase bound to indan | | Descriptor: | 1,2-ETHANEDIOL, 2,3-dihydro-1H-indene, FE (III) ION, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | One enzyme, many reactions: structural basis for the various reactions catalyzed by naphthalene 1,2-dioxygenase.
Iucrj, 4, 2017
|
|
4HM8
 
 | | Naphthalene 1,2-Dioxygenase bound to thioanisole | | Descriptor: | (methylsulfanyl)benzene, 1,2-ETHANEDIOL, FE (III) ION, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | One enzyme, many reactions: structural basis for the various reactions catalyzed by naphthalene 1,2-dioxygenase.
Iucrj, 4, 2017
|
|
4HM6
 
 | | Naphthalene 1,2-Dioxygenase bound to phenetole | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | One enzyme, many reactions: structural basis for the various reactions catalyzed by naphthalene 1,2-dioxygenase.
Iucrj, 4, 2017
|
|
2Z0X
 
 | | Crystal structure of ProX-CysSA complex from T. thermophilus | | Descriptor: | 5'-O-(N-(L-CYSTEINYL)-SULFAMOYL)ADENOSINE, Putative uncharacterized protein TTHA1699 | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of ProX-CysSA complex from T. thermophilus
To be Published
|
|
1WV9
 
 | | Crystal Structure of Rhodanese Homolog TT1651 from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 | | Descriptor: | Rhodanese Homolog TT1651 | | Authors: | Mizohata, E, Hattori, M, Tatsuguchi, A, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-12-12 | | Release date: | 2005-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the single-domain rhodanese homologue TTHA0613 from Thermus thermophilus HB8
Proteins, 64, 2006
|
|
1WNA
 
 | | Crystal structure of the hypothetical protein TT1805 from Thermus thermophillus HB8 | | Descriptor: | the hypothetical protein (TT1805) | | Authors: | Pioszak, A.A, Kishishita, S, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-28 | | Release date: | 2005-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of the hypothetical protein TT1805 from Thermus thermophillus HB8
To be Published
|
|
2Z11
 
 | | Crystal structure of putative acetyltransferase | | Descriptor: | Ribosomal-protein-alanine acetyltransferase | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of putative acetyltransferase
To be Published
|
|
1NZ8
 
 | | Solution Structure of the N-utilization substance G (NusG) N-terminal (NGN) domain from Thermus thermophilus | | Descriptor: | TRANSCRIPTION ANTITERMINATION PROTEIN NUSG | | Authors: | Reay, P, Yamasaki, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus
Proteins, 56, 2004
|
|
1WOT
 
 | | Structure of putative minimal nucleotidyltransferase | | Descriptor: | PUTATIVE MINIMAL NUCLEOTIDYLTRANSFERASE | | Authors: | Suzuki, S, Hatanaka, H, Hondoh, T, Okumura, A, Kuroda, Y, Kuramitsu, S, Shibata, T, Inoue, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-08-25 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of Putative Minimal Nucleotidyltransferase
To be Published
|
|
1WDT
 
 | | Crystal structure of ttk003000868 from Thermus thermophilus HB8 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, elongation factor G homolog | | Authors: | Wang, H, Takemoto-Hori, C, Murayama, K, Sekine, S, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-17 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of ttk003000868 from Thermus thermophilus HB8
To be published
|
|
3CBN
 
 | |
3EB2
 
 | |
