5ONJ
 
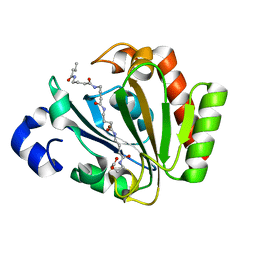 | | YnDL in Complex with 5 amino acid (PGA) complex | | Descriptor: | (2R,3S,4R,5R,6R)-6-((1R,2R,3S,4R,6S)-4,6-DIAMINO-2,3-DIHYDROXYCYCLOHEXYLOXY)-5-AMINO-2-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-3,4-DIOL, YndL, ZINC ION | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
Febs J., 285, 2018
|
|
5C6T
 
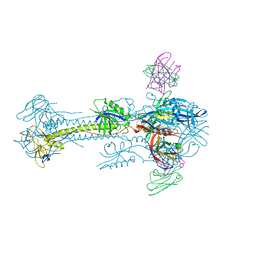 | | Crystal structure of HCMV glycoprotein B in complex with 1G2 Fab | | Descriptor: | 1G2 Fab heavy chain, 1G2 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chandramouli, S, Ciferri, C, Settembre, E.C, Carfi, A. | | Deposit date: | 2015-06-23 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of HCMV glycoprotein B in the postfusion conformation bound to a neutralizing human antibody.
Nat Commun, 6, 2015
|
|
6HRJ
 
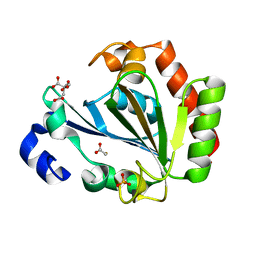 | | Apo - YndL | | Descriptor: | CITRATE ANION, SULFATE ION, YndL, ... | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2018-09-27 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
FEBS J., 285, 2018
|
|
5ONK
 
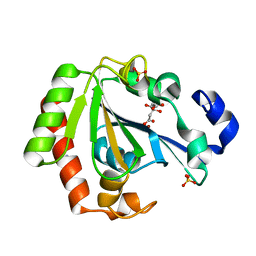 | | Native YndL | | Descriptor: | CITRATE ANION, IMIDAZOLE, SULFATE ION, ... | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
FEBS J., 285, 2018
|
|
6HRI
 
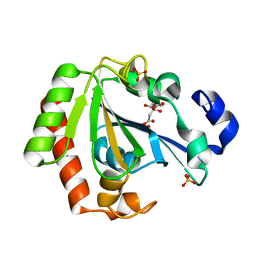 | | Native YndL | | Descriptor: | CITRATE ANION, IMIDAZOLE, SULFATE ION, ... | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2018-09-27 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
FEBS J., 285, 2018
|
|
5ONL
 
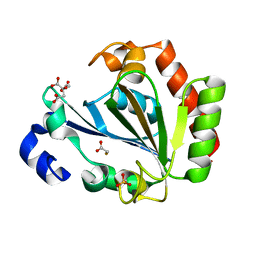 | | YNDL-apo (Zinc-free) | | Descriptor: | CITRATE ANION, SULFATE ION, YndL, ... | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
Febs J., 285, 2018
|
|
6AIE
 
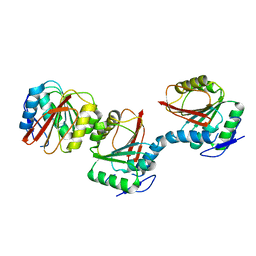 | | Crystal structure of a new form of RsmD-like RNA methyl transferase from Mycobacterium tuberculosis determined at 1.74 A resolution | | Descriptor: | Putative methyltransferase | | Authors: | Venkataraman, S, Dhankar, A, Sinha, K.M, Manivasakan, P, Iqbal, N, Singh, T.P, Prasad, B.V.L.S. | | Deposit date: | 2018-08-22 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of a new form of RsmD-like RNA methyl transferase from Mycobacterium tuberculosis determined at 1.74 A resolution
To Be Published
|
|
5XBP
 
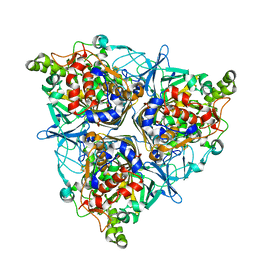 | | Oxygenase component of 3-nitrotoluene dioxygenase from Diaphorobacter sp. strain DS2 | | Descriptor: | 3NT oxygenase alpha subunit, 3NT oxygenase beta subunit, FE (III) ION, ... | | Authors: | Ramaswamy, S, Kumari, A, Singh, D, Gurunath, R. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional studies of ferredoxin and oxygenase components of 3-nitrotoluene dioxygenase from Diaphorobacter sp. strain DS2.
PLoS ONE, 12, 2017
|
|
5BOK
 
 | |
6A8T
 
 | |
5X8Z
 
 | |
5X9I
 
 | |
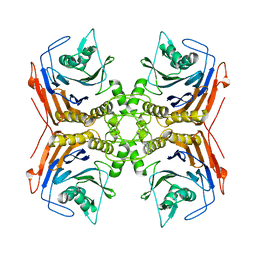
6A8Q
 
 | |
422D
 
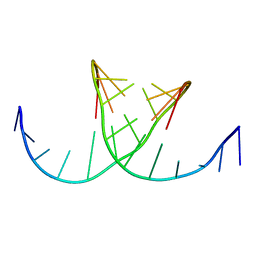 | |
2X12
 
 | |
4MMP
 
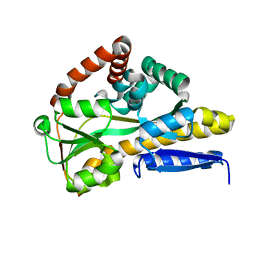 | |
4MYM
 
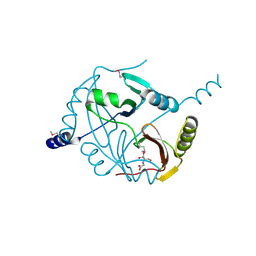 | | Crystal structure of a glyoxalase/ bleomycin resistance protein/ dioxygenase from Nocardioides | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glyoxalase/bleomycin resistance protein/dioxygenase | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, AL Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-09-27 | | Release date: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a glyoxalase/ bleomycin resistance protein/ dioxygenase from Nocardioides.
To be Published
|
|
4FGS
 
 | | Crystal structure of a probable dehydrogenase protein | | Descriptor: | Probable dehydrogenase protein, SULFATE ION | | Authors: | Eswaramoorthy, S, Rice, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-06-04 | | Release date: | 2012-08-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of a probable dehydrogenase protein
To be Published
|
|
4EW6
 
 | |
4J3C
 
 | | Crystal structure of 16S ribosomal RNA methyltransferase RsmE | | Descriptor: | Ribosomal RNA small subunit methyltransferase E | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-05 | | Release date: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 16S ribosomal RNA methyltransferase RsmE
TO BE PUBLISHED
|
|
4JJ9
 
 | | Crystal Structure of 5-carboxymethyl-2-hydroxymuconate delta-isomerase | | Descriptor: | 5-carboxymethyl-2-hydroxymuconate delta-isomerase, SULFATE ION | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-07 | | Release date: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of 5-carboxymethyl-2-hydroxymuconate delta-isomerase
To be Published
|
|
4JOT
 
 | | Crystal structure of enoyl-CoA hydrotase from Deinococcus radiodurans R1 | | Descriptor: | Enoyl-CoA hydratase, putative, GLYCEROL | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-18 | | Release date: | 2013-04-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of enoyl-CoA hydrotase from Deinococcus radiodurans R1
To be Published
|
|
4JCS
 
 | | Crystal structure of Enoyl-CoA hydratase/isomerase from Cupriavidus metallidurans CH34 | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Eswaramoorthy, S, Chamala, S, Chamala, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-22 | | Release date: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Enoyl-CoA hydratase/isomerase from Cupriavidus metallidurans CH34
To be Published
|
|
4K29
 
 | | Crystal structure of an enoyl-CoA hydratase/isomerase from Xanthobacter autotrophicus Py2 | | Descriptor: | Enoyl-CoA hydratase/isomerase, GLYCEROL, L(+)-TARTARIC ACID | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-08 | | Release date: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of an enoyl-CoA hydratase/isomerase from Xanthobacter autotrophicus Py2
TO BE PUBLISHED
|
|
4K2N
 
 | | Crystal structure of an enoyl-CoA hydratase/ carnithine racemase from Magnetospirillum magneticum | | Descriptor: | Enoyl-CoA hydratase/carnithine racemase | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-09 | | Release date: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an enoyl-CoA hydratase/ carnithine racemase from Magnetospirillum magneticum
To be Published
|
|
