3GX1
 
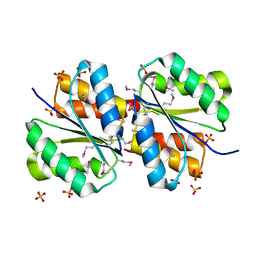 | |
3LSG
 
 | |
4K0D
 
 | | Periplasmic sensor domain of sensor histidine kinase, Adeh_2942 | | Descriptor: | ACETATE ION, CHLORIDE ION, Periplasmic sensor hybrid histidine kinase, ... | | Authors: | Pokkuluri, P.R, Mack, J.C, Bearden, J, Rakowski, E, Schiffer, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-04-03 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Analysis of periplasmic sensor domains from Anaeromyxobacter dehalogenans 2CP-C: structure of one sensor domain from a histidine kinase and another from a chemotaxis protein.
Microbiologyopen, 2, 2013
|
|
4K08
 
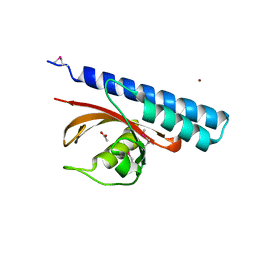 | | Periplasmic sensor domain of chemotaxis protein, Adeh_3718 | | Descriptor: | ACETATE ION, Methyl-accepting chemotaxis sensory transducer, ZINC ION | | Authors: | Pokkuluri, P.R, Mack, J.C, Bearden, J, Rakowski, E, Schiffer, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-04-03 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of periplasmic sensor domains from Anaeromyxobacter dehalogenans 2CP-C: structure of one sensor domain from a histidine kinase and another from a chemotaxis protein.
Microbiologyopen, 2, 2013
|
|
3MFD
 
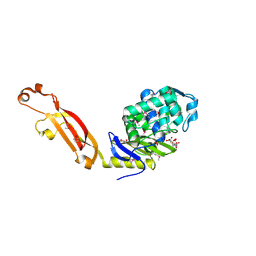 | | The Structure of the Beta-lactamase superfamily domain of D-alanyl-D-alanine carboxypeptidase from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, D-alanyl-D-alanine carboxypeptidase dacB | | Authors: | Cuff, M.E, Rakowski, E, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-01 | | Release date: | 2010-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Structure of the Beta-lactamase superfamily domain of D-alanyl-D-alanine carboxypeptidase from Bacillus subtilis.
TO BE PUBLISHED
|
|
3L1W
 
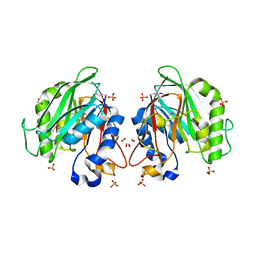 | | The crystal structure of a functionally unknown conserved protein from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, SULFATE ION, ... | | Authors: | Tan, K, Rakowski, E, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein from Enterococcus faecalis V583
To be Published
|
|
3O60
 
 | |
3IQT
 
 | | Structure of the HPT domain of Sensor protein barA from Escherichia coli CFT073. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Signal transduction histidine-protein kinase barA | | Authors: | Cuff, M.E, Rakowski, E, Kim, Y, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-20 | | Release date: | 2009-09-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the HPT domain of Sensor protein barA from Escherichia coli CFT073.
TO BE PUBLISHED
|
|
3M0Z
 
 | | Crystal structure of putative aldolase from Klebsiella pneumoniae. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Chang, C, Rakowski, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of putative aldolase from Klebsiella pneumoniae.
To be Published
|
|
3IC9
 
 | | The structure of dihydrolipoamide dehydrogenase from Colwellia psychrerythraea 34H. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SODIUM ION, dihydrolipoamide dehydrogenase | | Authors: | Tan, K, Rakowski, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of dihydrolipoamide dehydrogenase from Colwellia psychrerythraea 34H.
To be Published
|
|
3IR9
 
 | |
3KMI
 
 | | Crystal structure of putative membrane protein from Clostridium difficile 630 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Putative membrane protein COG4129 | | Authors: | Chang, C, Rakowski, E, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-11-10 | | Release date: | 2009-11-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of putative membrane protein from Clostridium difficile 630
To be Published
|
|
3NE8
 
 | | The crystal structure of a domain from N-acetylmuramoyl-l-alanine amidase of Bartonella henselae str. Houston-1 | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Rakowski, E, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-08 | | Release date: | 2010-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.239 Å) | | Cite: | A conformational switch controls cell wall-remodelling enzymes required for bacterial cell division.
Mol.Microbiol., 85, 2012
|
|
