5CZL
 
 | | Crystal structure of a novel GH8 endo-beta-1,4-glucanase from an Achatina fulica gut metagenomic library | | Descriptor: | Glucanase, PHOSPHATE ION | | Authors: | Scapin, S.M.N, Souza, F.H.M, Zanphorlin, L.M, Almeida, T.S, Sade, Y.B, Cardoso, A.M, Pinheiro, G.L, Murakami, M.T. | | Deposit date: | 2015-07-31 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Crystal structure of a novel GH8 endo-beta-1,4-glucanase from an Achatina fulica gut metagenomic library
To Be Published
|
|
4KC7
 
 | | Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Glycoside hydrolase, ... | | Authors: | Nascimento, A.F.Z, Polo, C.C, Santos, C.R, Costa, M.C.M.F, Mesa, A.N, Prade, R.A, Ruller, R, Squina, F.M, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
5DOM
 
 | | Crystal structure, maturation and flocculating properties of a 2S albumin from Moringa oleifera seeds | | Descriptor: | 1,2-ETHANEDIOL, 2S albumin, ACETATE ION | | Authors: | Ullah, A, Murakami, M.T, Arni, R.K. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-11 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of mature 2S albumin from Moringa oleifera seeds.
Biochem.Biophys.Res.Commun., 468, 2015
|
|
4MDO
 
 | | Crystal structure of a GH1 beta-glucosidase from the fungus Humicola insolens | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Giuseppe, P.O, Souza, T.A.C.B, Souza, F.H.M, Zanphorlin, L.M, Machado, C.B, Ward, R.J, Jorge, J.A, Furriel, R.P.M, Murakami, M.T. | | Deposit date: | 2013-08-23 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for glucose tolerance in GH1 beta-glucosidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3PZT
 
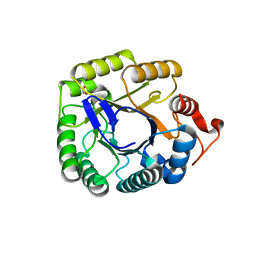 | | Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion | | Descriptor: | Endoglucanase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Santos, C.R, Paiva, J.H, Akao, P.K, Meza, A.N, Silva, J.C, Squina, F.M, Ward, R.J, Ruller, R, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Dissecting structure-function-stability relationships of a thermostable GH5-CBM3 cellulase from Bacillus subtilis 168.
Biochem.J., 441, 2012
|
|
1A2C
 
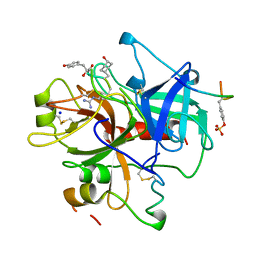 | | Structure of thrombin inhibited by AERUGINOSIN298-A from a BLUE-GREEN ALGA | | Descriptor: | Aeruginosin 298-A, Hirudin variant-2, SODIUM ION, ... | | Authors: | Rios-Steiner, J.L, Murakami, M, Tulinsky, A. | | Deposit date: | 1997-12-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Thrombin Inhibited by Aeruginosin 298-A from a Blue-Green Alga
J.Am.Chem.Soc., 120, 1998
|
|
2ROG
 
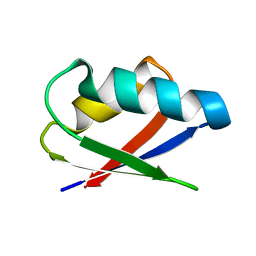 | | Solution structure of Thermus thermophilus HB8 TTHA1718 protein in living E. coli cells | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-21 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
6N98
 
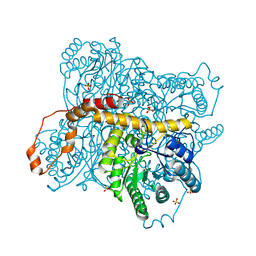 | | Xylose isomerase 1F1 variant from Streptomyces sp. F-1 | | Descriptor: | MAGNESIUM ION, SULFATE ION, Xylose isomerase | | Authors: | Miyamoto, R.Y, Vieira, P.S, Murakami, M.T, Zanphorlin, L.M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of a novel xylose isomerase from Streptomyces sp. F-1 revealed the presence of unique features that differ from conventional classes.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
4KCA
 
 | | Crystal Structure of Endo-1,5-alpha-L-arabinanase from a Bovine Ruminal Metagenomic Library | | Descriptor: | Endo-1,5-alpha-L-arabinanase, GLYCEROL, IODIDE ION, ... | | Authors: | Santos, C.R, Polo, C.C, Costa, M.C.M.F, Nascimento, A.F.Z, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2014-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
4GV5
 
 | | X-ray structure of crotamine, a cell-penetrating peptide from the Brazilian snake Crotalus durissus terrificus | | Descriptor: | Crotamine Ile-19, GLYCEROL, SULFATE ION, ... | | Authors: | Coronado, M.A, Gabdulkhakov, A, Georgieva, D, Sankaran, B, Murakami, M.T, Arni, R.K, Betzel, C. | | Deposit date: | 2012-08-30 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the polypeptide crotamine from the Brazilian rattlesnake Crotalus durissus terrificus.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2ROE
 
 | | Solution structure of thermus thermophilus HB8 TTHA1718 protein in vitro | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-20 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
4M1R
 
 | | Structure of a novel cellulase 5 from a sugarcane soil metagenomic library | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellulase 5 | | Authors: | Paiva, J.H, Alvarez, T.M, Cairo, J.P, Paixao, D.A, Almeida, R.A, Tonoli, C.C.C, Ruiz, D.M, Ruller, R, Santos, C.R, Squina, F.M, Murakami, M.T. | | Deposit date: | 2013-08-03 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of a novel cellulase 5 from sugarcane soil metagenome.
Plos One, 8, 2013
|
|
1YM4
 
 | | Crystal structure of human beta secretase complexed with NVP-AMK640 | | Descriptor: | Beta-secretase 1, NVP-AMK640 INHIBITOR | | Authors: | Hanessian, S, Yun, H, Hou, Y, Yang, G, Bayrakdarian, M, Therrien, E, Moitessier, N, Roggo, S, Veenstra, S. | | Deposit date: | 2005-01-20 | | Release date: | 2006-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based design, synthesis, and memapsin 2 (BACE) inhibitory activity of carbocyclic and heterocyclic peptidomimetics
J.Med.Chem., 48, 2005
|
|
1YM2
 
 | | Crystal structure of human beta secretase complexed with NVP-AUR200 | | Descriptor: | Beta-secretase 1, NVP-AUR200 INHIBITOR | | Authors: | Hanessian, S, Yun, H, Hou, Y, Yang, G, Bayrakdarian, M, Therrien, E, Moitessier, N, Roggo, S, Veenstra, S. | | Deposit date: | 2005-01-20 | | Release date: | 2006-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-based design, synthesis, and memapsin 2 (BACE) inhibitory activity of carbocyclic and heterocyclic peptidomimetics
J.Med.Chem., 48, 2005
|
|
2L8A
 
 | | Structure of a novel CBM3 lacking the calcium-binding site | | Descriptor: | Endoglucanase | | Authors: | Paiva, J.H, Meza, A.N, Sforca, M.L, Navarro, R.Z, Neves, J.L, Santos, C.R, Murakami, M.T, Zeri, A.C. | | Deposit date: | 2011-01-07 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dissecting structure-function-stability relationships of a thermostable GH5-CBM3 cellulase from Bacillus subtilis 168.
Biochem.J., 441, 2012
|
|
1WT7
 
 | | Solution structure of BuTX-MTX: a butantoxin-maurotoxin chimera | | Descriptor: | BuTX-MTX | | Authors: | M'Barek, S, Chagot, B, Andreotti, N, Visan, V, Mansuelle, P, Grissmer, S, Marrakchi, M, El Ayeb, M, Sampieri, F, Darbon, H, Fajloun, Z, De Waard, M, Sabatier, J.-M. | | Deposit date: | 2004-11-16 | | Release date: | 2004-11-30 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Increasing the molecular contacts between maurotoxin and Kv1.2 channel augments ligand affinity.
Proteins, 60, 2005
|
|
4KC8
 
 | | Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 in complex with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Glycoside hydrolase, ... | | Authors: | Nascimento, A.F.Z, Polo, C.C, Santos, C.R, Costa, M.C.M.F, Mesa, A.N, Prade, R.A, Ruller, R, Squina, F.M, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
4K68
 
 | | Structure of a novel GH10 endoxylanase retrieved from sugarcane soil metagenome | | Descriptor: | GH10 xylanase, GLYCEROL | | Authors: | Santos, C.R, Polo, C.C, Alvarez, T.M, Paixao, D.A.A, Almeida, R.F, Pereira, I.O, Squina, F.M, Murakami, M.T. | | Deposit date: | 2013-04-15 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Development and biotechnological application of a novel endoxylanase family GH10 identified from sugarcane soil metagenome.
Plos One, 8, 2013
|
|
4KCB
 
 | | Crystal Structure of Exo-1,5-alpha-L-arabinanase from Bovine Ruminal Metagenomic Library | | Descriptor: | Arabinan endo-1,5-alpha-L-arabinosidase, PHOSPHATE ION | | Authors: | Santos, C.R, Polo, C.C, Costa, M.C.M.F, Nascimento, A.F.Z, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2014-04-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
5DT5
 
 | | Crystal structure of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7 in space group P21 | | Descriptor: | Beta-glucosidase, SULFATE ION | | Authors: | Zanphorlin, L.M, Giuseppe, P.O, Tonoli, C.C.C, Murakami, M.T. | | Deposit date: | 2015-09-17 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Oligomerization as a strategy for cold adaptation: Structure and dynamics of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7.
Sci Rep, 6, 2016
|
|
4EWL
 
 | | Crystal Structure of MshB with glycerol and Acetate bound in the active site | | Descriptor: | 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside deacetylase, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ACETATE ION, ... | | Authors: | Broadley, S.G, Sewell, B.T, Weber, B.W, Marakalala, M.J, Steenkamp, D.J. | | Deposit date: | 2012-04-27 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A new crystal form of MshB from Mycobacterium tuberculosis with glycerol and acetate in the active site suggests the catalytic mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4NPR
 
 | | Crystal Structure of the Family 12 Xyloglucanase from Aspergillus niveus | | Descriptor: | SULFATE ION, Xyloglucan-specific endo-beta-1,4-glucanase GH12 | | Authors: | Cordeiro, R.L, Santos, C.R, Furtado, G.P, Damasio, A.R.L, Polizeli, M.L.T.M, Ward, R.J, Murakami, M.T. | | Deposit date: | 2013-11-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Family 12 Xyloglucanase from Aspergillus niveus
To be Published
|
|
1AQ7
 
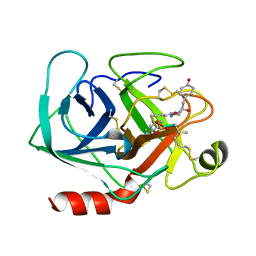 | |
1B9O
 
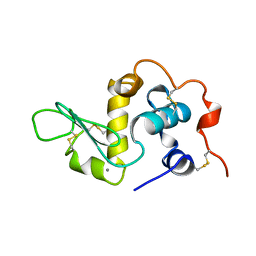 | | HUMAN ALPHA-LACTALBUMIN, LOW TEMPERATURE FORM | | Descriptor: | CALCIUM ION, PROTEIN (ALPHA-LACTALBUMIN) | | Authors: | Harata, K, Abe, Y, Muraki, M. | | Deposit date: | 1999-02-14 | | Release date: | 1999-03-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystallographic evaluation of internal motion of human alpha-lactalbumin refined by full-matrix least-squares method.
J.Mol.Biol., 287, 1999
|
|
5DT7
 
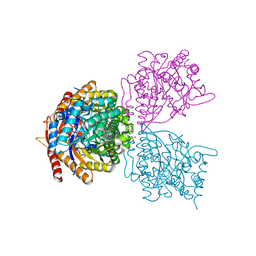 | | Crystal structure of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7 in space group C2221 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Zanphorlin, L.M, Giuseppe, P.O, Tonoli, C.C.C, Murakami, M.T. | | Deposit date: | 2015-09-17 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Oligomerization as a strategy for cold adaptation: Structure and dynamics of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7.
Sci Rep, 6, 2016
|
|
