1QGA
 
 | | PEA FNR Y308W MUTANT IN COMPLEX WITH NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE), ... | | Authors: | Deng, Z, Aliverti, A, Zanetti, G, Arakaki, A.K, Ottado, J, Orellano, E.G, Calcaterra, N.B, Ceccarelli, E.A, Carrillo, N, Karplus, P.A. | | Deposit date: | 1999-04-18 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A productive NADP+ binding mode of ferredoxin-NADP+ reductase revealed by protein engineering and crystallographic studies.
Nat.Struct.Biol., 6, 1999
|
|
1QIU
 
 | |
5WG8
 
 | | Structure of PP5C with LB-100; 7-oxabicyclo[2.2.1]heptane-2,3-dicarbonyl moiety modeled in the density | | Descriptor: | (1S,2R,3S,4R)-3-(4-methylpiperazine-1-carbonyl)-7-oxabicyclo[2.2.1]heptane-2-carboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | D'Arcy, B.M, Swingle, M.R, Honkanen, R.E, Prakash, A. | | Deposit date: | 2017-07-13 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Antitumor Drug LB-100 Is a Catalytic Inhibitor of Protein Phosphatase 2A (PPP2CA) and 5 (PPP5C) Coordinating with the Active-Site Catalytic Metals in PPP5C.
Mol. Cancer Ther., 18, 2019
|
|
6YVE
 
 | | Glycogen phosphorylase b in complex with pelargonidin 3-O-beta-D-glucoside | | Descriptor: | DIMETHYL SULFOXIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Drakou, C.E, Gardeli, C, Tsialtas, I, Alexopoulos, S, Mallouchos, A, Koulas, S, Tsagkarakou, A, Asimakopoulos, D, Leonidas, D.D, Psarra, A.M, Skamnaki, V.T. | | Deposit date: | 2020-04-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Affinity Crystallography Reveals Binding of Pomegranate Juice Anthocyanins at the Inhibitor Site of Glycogen Phosphorylase: The Contribution of a Sugar Moiety to Potency and Its Implications to the Binding Mode.
J.Agric.Food Chem., 68, 2020
|
|
7P0K
 
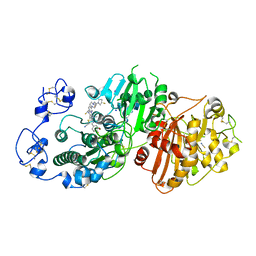 | | Crystal structure of Autotaxin (ENPP2) with 18F-labeled positron emission tomography ligand | | Descriptor: | 2-[[2-ethyl-6-[4-[2-[(3~{R})-3-fluoranylpyrrolidin-1-yl]-2-oxidanylidene-ethyl]piperazin-1-yl]imidazo[1,2-a]pyridin-3-yl]-methyl-amino]-4-(4-fluorophenyl)-2,3-dihydro-1,3-thiazole-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salgado-Polo, F, Shao, T, Xiao, Z, Van, R, Chen, J, Rong, J, Haider, A, Shao, Y, Josephson, L, Perrakis, A, Liang, S.H. | | Deposit date: | 2021-06-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Imaging Autotaxin In Vivo with 18 F-Labeled Positron Emission Tomography Ligands
J Med Chem, 64, 2021
|
|
9GSO
 
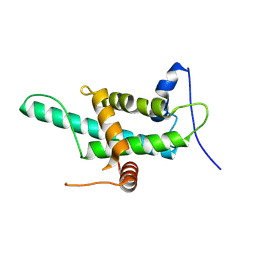 | |
9EPP
 
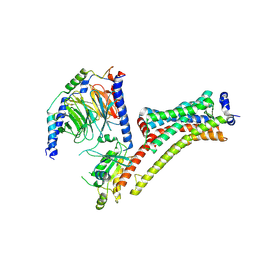 | | Cryo-EM Structure of Jumping Spider Rhodopsin-1 bound to a Giq heterotrimer | | Descriptor: | 11,20-Ethanoretinal, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tejero, O, Pamula, F, Koyanagi, M, Nagata, T, Afanasyev, P, Das, I, Deupi, X, Sheves, M, Terakita, A, Schertler, G.F.X, Rodrigues, M.J, Tsai, C.-J. | | Deposit date: | 2024-03-19 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Active state structures of a bistable visual opsin bound to G proteins.
Nat Commun, 15, 2024
|
|
9GSQ
 
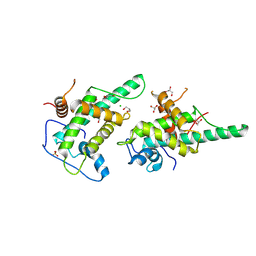 | | DNA binding domain of J-DNA Binding Protein 3 (JBP3) | | Descriptor: | CHLORIDE ION, DNA binding domain of J-DNA binding protein 3, GLYCEROL, ... | | Authors: | de Vries, I, Adamopoulos, A, Joosten, R.P, Perrakis, A. | | Deposit date: | 2024-09-16 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Base-J binding proteins JBP1 and JBP3 have conserved structures of their J-DNA binding domain but drastically different affinities
To Be Published
|
|
9EPQ
 
 | | Cryo-EM Structure of Jumping Spider Rhodopsin-1 bound to a Giq heterotrimer | | Descriptor: | 11,20-Ethanoretinal, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tejero, O, Pamula, F, Koyanagi, M, Nagata, T, Afanasyev, P, Das, I, Deupi, X, Sheves, M, Terakita, A, Schertler, G.F.X, Rodrigues, M.J, Tsai, C.-J. | | Deposit date: | 2024-03-19 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Active state structures of a bistable visual opsin bound to G proteins.
Nat Commun, 15, 2024
|
|
8BBM
 
 | |
6FPV
 
 | | A llama-derived JBP1-targeting nanobody | | Descriptor: | GLYCEROL, Nanobody | | Authors: | van Beusekom, B, Adamopoulos, A, Heidebrecht, T, Joosten, R.P, Perrakis, A. | | Deposit date: | 2018-02-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Characterization and structure determination of a llama-derived nanobody targeting the J-base binding protein 1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6NVQ
 
 | |
7ZU3
 
 | |
7ZUA
 
 | |
8A2E
 
 | | Crystal Structure of Human Parechovirus 3 2A protein | | Descriptor: | 2A protein, GLYCEROL, SULFATE ION | | Authors: | von Castelmur, E, Zhu, L, wang, X, Fry, E, Ren, J, Perrakis, A, Stuart, D.I. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural plasticity of 2A proteins in the Parechovirus family.
To Be Published
|
|
6GVJ
 
 | | Human Mps1 kinase domain with ordered activation loop | | Descriptor: | CHLORIDE ION, Dual specificity protein kinase TTK, GLYCEROL | | Authors: | Roorda, J.C, Hiruma, Y, Joosten, R.P, Perrakis, A. | | Deposit date: | 2018-06-21 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A crystal structure of the human protein kinase Mps1 reveals an ordered conformation of the activation loop.
Proteins, 87, 2019
|
|
8E10
 
 | | Structure of mouse polymerase beta | | Descriptor: | DNA polymerase beta, MALONIC ACID | | Authors: | Thompson, M.K, Sharma, N, Prakash, A. | | Deposit date: | 2022-08-09 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Pol beta /XRCC1 heterodimerization dictates DNA damage recognition and basal Pol beta protein levels without interfering with mouse viability or fertility.
DNA Repair (Amst), 123, 2023
|
|
8E11
 
 | | Structure of mouse DNA polymerase Beta (PolB) mutant | | Descriptor: | ACETATE ION, DNA polymerase beta, MALONIC ACID | | Authors: | Sharma, N, Thompson, M.K, Prakash, A. | | Deposit date: | 2022-08-09 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pol beta /XRCC1 heterodimerization dictates DNA damage recognition and basal Pol beta protein levels without interfering with mouse viability or fertility.
DNA Repair (Amst), 123, 2023
|
|
4WEB
 
 | | Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FORMAMIDE, Mouse Fab Heavy Chain, ... | | Authors: | Khan, A.G, Whidby, J, Miller, M.T, Scarborough, H, Zatorski, A.V, Cygan, A, Price, A.A, Yost, S.A, Bohannon, C.D, Jacob, J, Grakoui, A, Marcotrigiano, J. | | Deposit date: | 2014-09-09 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2.
Nature, 509, 2014
|
|
7ZUO
 
 | |
7ZU4
 
 | |
8A2F
 
 | | Crystal Structure of Ljunganvirus 1 2A protein | | Descriptor: | 2A protein | | Authors: | von Castelmur, E, Zhu, L, Wang, X, Fry, E, Ren, J, Perrakis, A, Stuart, D.I. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural plasticity of 2A proteins in the Parechovirus family
To Be Published
|
|
5LJJ
 
 | | Crystal structure of human Mps1 (TTK) in complex with Reversine | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase TTK, N~6~-cyclohexyl-N~2~-(4-morpholin-4-ylphenyl)-9H-purine-2,6-diamine | | Authors: | Hiruma, Y, Joosten, R.P, Perrakis, A. | | Deposit date: | 2016-07-18 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of reversine selectivity in inhibiting Mps1 more potently than aurora B kinase.
Proteins, 84, 2016
|
|
7ZOM
 
 | |
7ZV6
 
 | |
