3H98
 
 | | Crystal structure of HCV NS5b 1b with (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyrimidine derivative | | 分子名称: | GLYCEROL, N-{3-[5-hydroxy-8-(3-methylbutyl)-7-oxo-7,8-dihydroimidazo[1,2-a]pyrimidin-6-yl]-1,1-dioxido-4H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | 著者 | Wang, G, Lei, H, Wang, X, Das, D, Mackinnon, C, Montalbetti, C.A.G, Mears, R, Gai, X, Bailey, S, Ruhrmund, D, Hooi, L, Misialek, S, Rajagopalan, R, Cheng, R.K.Y, Barker, J.L, Felicetti, B, Stoycheva, A, Buckman, B, Kossen, K, Seiwert, S, Beigelmana, L. | | 登録日 | 2009-04-30 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | HCV NS5B polymerase inhibitors 2: Synthesis and in vitro activity of (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyridine and azolo[1,5-a]pyrimidine derivatives.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1SGU
 
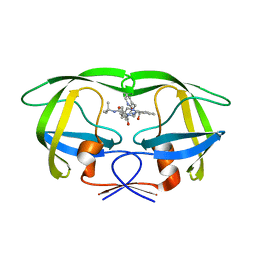 | | Comparing the Accumulation of Active Site and Non-active Site Mutations in the HIV-1 Protease | | 分子名称: | N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, POL polyprotein | | 著者 | Clemente, J.C, Moose, R.E, Hemrajani, R, Govindasamy, L, Reutzel, R, Mckenna, R, Abandje-McKenna, M, Goodenow, M.M, Dunn, B.M. | | 登録日 | 2004-02-24 | | 公開日 | 2004-10-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Comparing the Accumulation of Active- and Nonactive-Site Mutations in the HIV-1 Protease.
Biochemistry, 43, 2004
|
|
1SH9
 
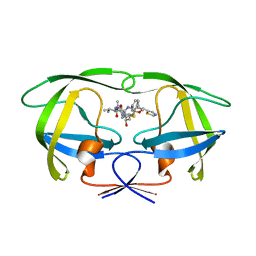 | | Comparing the Accumulation of Active Site and Non-active Site Mutations in the HIV-1 Protease | | 分子名称: | POL polyprotein, RITONAVIR | | 著者 | Clemente, J.C, Moose, R.E, Hemrajani, R, Govindasamy, L, Reutzel, R, McKenna, R, Abanje-McKenna, M, Goodenow, M.M, Dunn, B.M. | | 登録日 | 2004-02-25 | | 公開日 | 2004-10-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Comparing the Accumulation of Active- and Nonactive-Site Mutations in the HIV-1 Protease.
Biochemistry, 43, 2004
|
|
8WSV
 
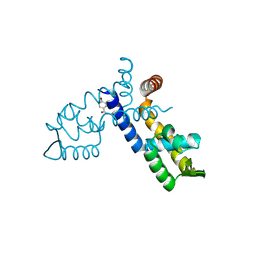 | |
8XB7
 
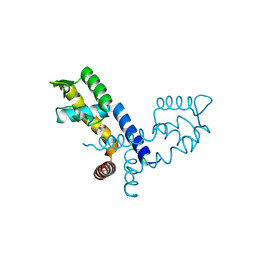 | |
5KR8
 
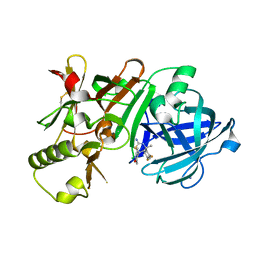 | | (4~{S},6~{S})-4-[2,4-bis(fluoranyl)phenyl]-6-(3,5-dimethyl-1,2-oxazol-4-yl)-4-methyl-5,6-dihydro-1,3-thiazin-2-amine (compound 5) bound to BACE1 | | 分子名称: | (4~{S},6~{S})-4-[2,4-bis(fluoranyl)phenyl]-6-(3,5-dimethyl-1,2-oxazol-4-yl)-4-methyl-5,6-dihydro-1,3-thiazin-2-amine, Beta-secretase 1, IODIDE ION | | 著者 | Lewis, H.A, Wu, Y.J, Rajamani, R, Thompson, L.A. | | 登録日 | 2016-07-07 | | 公開日 | 2016-09-07 | | 最終更新日 | 2016-10-05 | | 実験手法 | X-RAY DIFFRACTION (2.118 Å) | | 主引用文献 | Discovery of S3-Truncated, C-6 Heteroaryl Substituted Aminothiazine beta-Site APP Cleaving Enzyme-1 (BACE1) Inhibitors.
J.Med.Chem., 59, 2016
|
|
1D5D
 
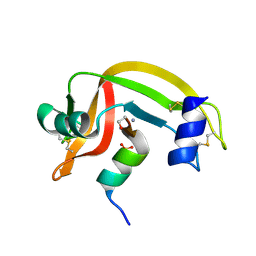 | |
1D5H
 
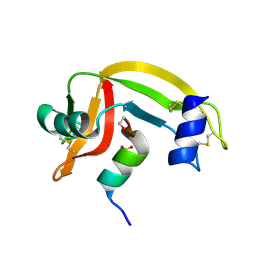 | | Rnase s(f8a). mutant ribonucleasE S. | | 分子名称: | RNASE S, S PEPTIDE, SULFATE ION | | 著者 | Ratnaparkhi, G.S, Varadarajan, R. | | 登録日 | 1999-10-07 | | 公開日 | 1999-10-20 | | 最終更新日 | 2018-03-14 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Thermodynamic and structural studies of cavity formation in proteins suggest that loss of packing interactions rather than the hydrophobic effect dominates the observed energetics.
Biochemistry, 39, 2000
|
|
2RLN
 
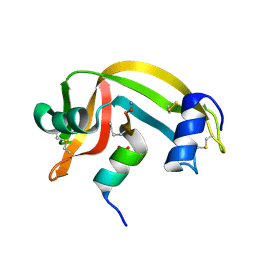 | |
4ZNP
 
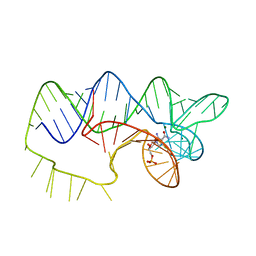 | | The structure of A pfI Riboswitch Bound to ZMP | | 分子名称: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, MAGNESIUM ION, pfI Riboswitch | | 著者 | Ren, A, Patel, D.J, Rajashankar, R.K. | | 登録日 | 2015-05-05 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Global RNA Fold and Molecular Recognition for a pfl Riboswitch Bound to ZMP, a Master Regulator of One-Carbon Metabolism.
Structure, 23, 2015
|
|
1J80
 
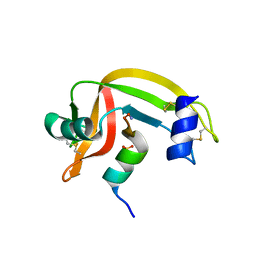 | | Osmolyte Stabilization of RNase | | 分子名称: | RIBONUCLEASE PANCREATIC, SULFATE ION | | 著者 | Ratnaparkhi, G.S, Varadarajan, R. | | 登録日 | 2001-05-19 | | 公開日 | 2001-06-06 | | 最終更新日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Osmolytes stabilize ribonuclease S by stabilizing its fragments S protein and S peptide to compact folding-competent states.
J.Biol.Chem., 276, 2001
|
|
1A19
 
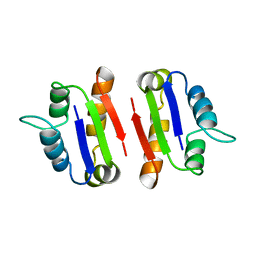 | | BARSTAR (FREE), C82A MUTANT | | 分子名称: | BARSTAR | | 著者 | Ratnaparkhi, G.S, Varadarajan, R. | | 登録日 | 1997-12-25 | | 公開日 | 1998-04-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Discrepancies between the NMR and X-ray structures of uncomplexed barstar: analysis suggests that packing densities of protein structures determined by NMR are unreliable.
Biochemistry, 37, 1998
|
|
1J82
 
 | | Osmolyte Stabilization of RNase | | 分子名称: | RIBONUCLEASE PANCREATIC, SULFATE ION | | 著者 | Ratnaparkhi, G.S, Varadarajan, R. | | 登録日 | 2001-05-19 | | 公開日 | 2001-06-06 | | 最終更新日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Osmolytes stabilize ribonuclease S by stabilizing its fragments S protein and S peptide to compact folding-competent states.
J.Biol.Chem., 276, 2001
|
|
1NKG
 
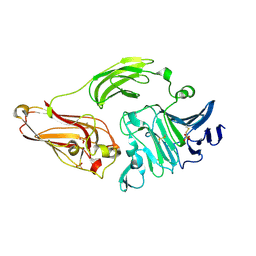 | | Rhamnogalacturonan lyase from Aspergillus aculeatus | | 分子名称: | CALCIUM ION, Rhamnogalacturonase B, SULFATE ION | | 著者 | McDonough, M.A, Kadirvelraj, R, Harris, P, Poulsen, J.C, Larsen, S. | | 登録日 | 2003-01-03 | | 公開日 | 2004-05-25 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Rhamnogalacturonan lyase reveals a unique three-domain modular structure for polysaccharide lyase family 4.
Febs Lett., 565, 2004
|
|
1J7Z
 
 | |
1J81
 
 | | Osmolyte Stabilization of RNase | | 分子名称: | RIBONUCLEASE PANCREATIC, SULFATE ION | | 著者 | Ratnaparkhi, G.S, Varadarajan, R. | | 登録日 | 2001-05-19 | | 公開日 | 2001-06-06 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Osmolytes stabilize ribonuclease S by stabilizing its fragments S protein and S peptide to compact folding-competent states.
J.Biol.Chem., 276, 2001
|
|
1D5E
 
 | |
1FEV
 
 | |
1CJQ
 
 | |
1XP8
 
 | |
5VCR
 
 | | Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase with bound uranium dioxide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase, CHLORIDE ION, ... | | 著者 | Sanders, J.H, Kadirvelraj, R, Wood, Z.A. | | 登録日 | 2017-03-31 | | 公開日 | 2018-04-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.992 Å) | | 主引用文献 | HumanN-acetylglucosaminyltransferase II substrate recognition uses a modular architecture that includes a convergent exosite.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1CJR
 
 | |
2RNS
 
 | | REFINEMENT OF THE CRYSTAL STRUCTURE OF RIBONUCLEASE S. COMPARISON WITH AND BETWEEN THE VARIOUS RIBONUCLEASE A STRUCTURES | | 分子名称: | RIBONUCLEASE S, SULFATE ION | | 著者 | Kim, E.E, Varadarajan, R, Wyckoff, H.W, Richards, F.M. | | 登録日 | 1992-02-19 | | 公開日 | 1994-01-31 | | 最終更新日 | 2019-08-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Refinement of the crystal structure of ribonuclease S. Comparison with and between the various ribonuclease A structures.
Biochemistry, 31, 1992
|
|
5VCM
 
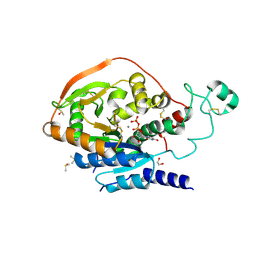 | | Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase with bound UDP and Manganese | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase, ... | | 著者 | Sanders, J.H, Kadirvelraj, R, Wood, Z.A. | | 登録日 | 2017-03-31 | | 公開日 | 2018-04-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | HumanN-acetylglucosaminyltransferase II substrate recognition uses a modular architecture that includes a convergent exosite.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3CWG
 
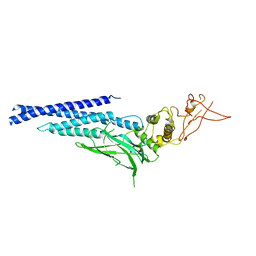 | | Unphosphorylated mouse STAT3 core fragment | | 分子名称: | Signal transducer and activator of transcription 3 | | 著者 | Ren, Z, Mao, X, Mertens, C, Krishnaraj, R, Qin, J, Mandal, P.K, Romanowshi, M.J, McMurray, J.S. | | 登録日 | 2008-04-21 | | 公開日 | 2008-07-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Crystal structure of unphosphorylated STAT3 core fragment.
Biochem.Biophys.Res.Commun., 374, 2008
|
|
