6HB8
 
 | | Crystal structure of OXA-517 beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, 2-ETHOXYETHANOL, Beta-lactamase, ... | | Authors: | Raczynska, J.E, Dabos, L, Zavala, A, Retailleau, P, Iorga, B, Jaskolski, M, Naas, T. | | Deposit date: | 2018-08-09 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Genetic, biochemical and structural characterization of OXA-517, an OXA-48-like extended-spectrum cephalosporins and carbapenems-hydrolyzing beta-lactamase
To Be Published
|
|
6OL8
 
 | | Crystal structure of NDM-12 metallo-beta-lactamase in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, Metallo-beta-lactamase NDM-12, ... | | Authors: | Raczynska, J.E, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Flexible loops of New Delhi metallo-beta-lactamase modulate its activity towards different substrates.
Int.J.Biol.Macromol., 158, 2020
|
|
6OGO
 
 | | Crystal structure of NDM-9 metallo-beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Raczynska, J.E, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Flexible loops of New Delhi metallo-beta-lactamase modulate its activity towards different substrates.
Int.J.Biol.Macromol., 158, 2020
|
|
5N0H
 
 | | Crystal structure of NDM-1 in complex with hydrolyzed meropenem - new refinement | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, King, D.T, Strynadka, N.C.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5N0I
 
 | | Crystal structure of NDM-1 in complex with beta-mercaptoethanol - new refinement | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, King, D.T, Strynadka, N.C.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5NBK
 
 | | NDM-1 metallo-beta-lactamase: a parsimonious interpretation of the diffraction data | | Descriptor: | CHLORIDE ION, HEXAETHYLENE GLYCOL, Metallo-beta-lactamase type 2, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-03-02 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5O2E
 
 | | Crystal structure of NDM-1 in complex with hydrolyzed cefuroxime - new refinement | | Descriptor: | (2R,5S)-5-[(carbamoyloxy)methyl]-2-[(R)-carboxy{[(2Z)-2-(furan-2-yl)-2-(methoxyimino)acetyl]amino}methyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Metallo-beta-lactamase type 2, SULFATE ION, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-05-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5O2F
 
 | | Crystal structure of NDM-1 in complex with hydrolyzed ampicillin - new refinement | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-05-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5W8W
 
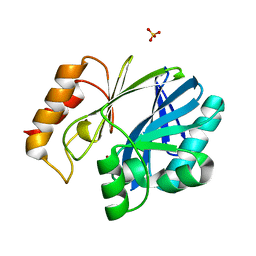 | | Bacillus cereus Zn-dependent metallo-beta-lactamase at pH 7 - new refinement | | Descriptor: | Metallo-beta-lactamase type 2, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Gonzalez, J.M, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Wlodawer, A, Gonzalez, M.M, Vila, A.J. | | Deposit date: | 2017-06-22 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
6EX7
 
 | | Crystal structure of NDM-1 metallo-beta-lactamase in complex with Cd ions and a hydrolyzed beta-lactam ligand - new refinement | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, CADMIUM ION, ... | | Authors: | Kim, Y, Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A. | | Deposit date: | 2017-11-07 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5WCM
 
 | | Crystal structure of the complex between class B3 beta-lactamase BJP-1 and 4-nitrobenzene-sulfonamide - new refinement | | Descriptor: | 4-nitrobenzenesulfonamide, Blr6230 protein, ZINC ION | | Authors: | Docquier, J.D, Benvenuti, M, Calderone, V, Menciassi, N, Shabalin, I.G, Raczynska, J.E, Wlodawer, A, Jaskolski, M, Minor, W, Mangani, S. | | Deposit date: | 2017-06-30 | | Release date: | 2017-07-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structure of the subclass B3 metallo-beta-lactamase BJP-1: rational basis for substrate specificity and interaction with sulfonamides.
Antimicrob. Agents Chemother., 54, 2010
|
|
4HMD
 
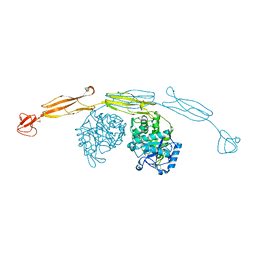 | | Crystal structure of cold-adapted chitinase from Moritella marina with a reaction intermediate - oxazolinium ion (NGO) | | Descriptor: | 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY-ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3-OXAZOLE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HME
 
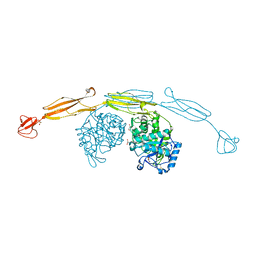 | | Crystal structure of cold-adapted chitinase from Moritella marina with a reaction product - NAG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HMC
 
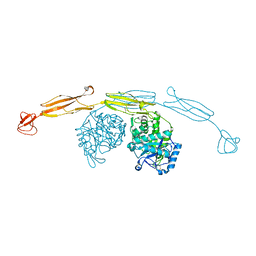 | | Crystal structure of cold-adapted chitinase from Moritella marina | | Descriptor: | Chitinase 60, GLYCEROL, SODIUM ION | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5W90
 
 | | FEZ-1 metallo-beta-lactamase from Legionella gormanii modelled with unknown ligand | | Descriptor: | FEZ-1 protein, GLYCEROL, SULFATE ION, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Kahn, R, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Papamicael, C, Frere, J.M, Galleni, M, Dideberg, O. | | Deposit date: | 2017-06-22 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-beta-lactamase from Fluoribacter gormanii, in native form and in complex with D-captopril.
J. Mol. Biol., 325, 2003
|
|
5WCK
 
 | | Native FEZ-1 metallo-beta-lactamase from Legionella gormanii | | Descriptor: | FEZ-1 protein, GLYCEROL, SULFATE ION, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Kahn, R, Papamicael, C, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Frere, J.M, Galleni, M, Dideberg, O. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-beta-lactamase from Fluoribacter gormanii, in native form and in complex with D-captopril.
J. Mol. Biol., 325, 2003
|
|
