6ENZ
 
 | | Crystal structure of mouse GADL1 | | Descriptor: | Acidic amino acid decarboxylase GADL1, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Raasakka, A, Mahootchi, E, Winge, I, Luan, W, Kursula, P, Haavik, J. | | Deposit date: | 2017-10-07 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the mouse acidic amino acid decarboxylase GADL1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6TU2
 
 | | Crystal structure of rat annexin A11 | | Descriptor: | Annexin, CALCIUM ION | | Authors: | Raasakka, A, Lillebostad, P, Vedeler, A, Kursula, P. | | Deposit date: | 2020-01-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the ALS Mutation Target Annexin A11 Reveals a Stabilising N-Terminal Segment.
Biomolecules, 10, 2020
|
|
7YWD
 
 | | Human GDAP1 core domain, trigonal crystal form | | Descriptor: | Ganglioside-induced differentiation-associated protein 1 | | Authors: | Raasakka, A, Kursula, P. | | Deposit date: | 2022-02-13 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into Charcot-Marie-Tooth disease-linked mutations in human GDAP1.
Febs Open Bio, 12, 2022
|
|
6HYF
 
 | |
4WCC
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation P225G | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, ADENOSINE-2'-MONOPHOSPHATE, CHLORIDE ION | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4WDB
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation R307Q, complexed with 2'-AMP | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, ADENOSINE-2'-MONOPHOSPHATE, CHLORIDE ION | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
6ZEK
 
 | | Crystal structure of mouse CSAD | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Cysteine sulfinic acid decarboxylase, ... | | Authors: | Mahootchi, E, Raasakka, A, Haavik, J, Kursula, P. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and substrate specificity determinants of the taurine biosynthetic enzyme cysteine sulphinic acid decarboxylase.
J.Struct.Biol., 213, 2021
|
|
3ZBR
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation H230S, crystallized with NADP | | Descriptor: | 2', 3'-CYCLIC-NUCLEOTIDE 3'-PHOSPHODIESTERASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Myllykoski, M, Raasakka, A, Lehtimaki, M, Han, H, Kursula, P. | | Deposit date: | 2012-11-13 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Crystallographic Analysis of the Reaction Cycle of 2',3'-Cyclic Nucleotide 3'-Phosphodiesterase, a Unique Member of the 2H Phosphoesterase Family
J.Mol.Biol., 425, 2013
|
|
3ZBS
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation V321A, crystallized with 2'-AMPS | | Descriptor: | 2', 3'-CYCLIC NUCLEOTIDE 3'-PHOSPHODIESTERASE, 2'-O-(sulfidophosphinato)adenosine, ... | | Authors: | Myllykoski, M, Raasakka, A, Lehtimaki, M, Han, H, Kursula, P. | | Deposit date: | 2012-11-13 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystallographic Analysis of the Reaction Cycle of 2',3'-Cyclic Nucleotide 3'-Phosphodiesterase, a Unique Member of the 2H Phosphoesterase Family
J.Mol.Biol., 425, 2013
|
|
3ZBZ
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation V321A, crystallized with 2'-AMPS | | Descriptor: | 2', 3'-CYCLIC-NUCLEOTIDE 3'-PHOSPHODIESTERASE, 2'-O-(sulfidophosphinato)adenosine, ... | | Authors: | Myllykoski, M, Raasakka, A, Lehtimaki, M, Han, H, Kursula, P. | | Deposit date: | 2012-11-14 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Analysis of the Reaction Cycle of 2',3'-Cyclic Nucleotide 3'-Phosphodiesterase, a Unique Member of the 2H Phosphoesterase Family
J.Mol.Biol., 425, 2013
|
|
4WDF
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation V321A, complexed with 2',5'-ADP | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, ADENOSINE-2'-5'-DIPHOSPHATE | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4WDG
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation V321A, complexed with 2',5'-ADP | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, ADENOSINE-2'-5'-DIPHOSPHATE | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4WBL
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation F235A | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, CHLORIDE ION, GLYCEROL | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-03 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4WDD
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation T232A, complexed with citrate | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4WDH
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation Y168A | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, ACETIC ACID, CHLORIDE ION | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
7A0A
 
 | | Crystal structure of mouse CSAD in apo form | | Descriptor: | Cysteine sulfinic acid decarboxylase, SODIUM ION, SULFATE ION | | Authors: | Mahootchi, E, Raasakka, A, Haavik, J, Kursula, P. | | Deposit date: | 2020-08-07 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of cysteine sulphinic acid decarboxylase reveals structural determinants for substrate specificity of pyridoxal phosphate-dependent decarboxylases
To be published
|
|
7ALM
 
 | |
6TA0
 
 | | Human NAMPT in complex with nicotinic acid and phosphoribosyl pyrophosphate | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, GLYCEROL, NICOTINIC ACID, ... | | Authors: | Houry, D, Raasakka, A, Kursula, P, Ziegler, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification of structural determinants of NAMPT activity and substrate selectivity
To Be Published
|
|
8PIA
 
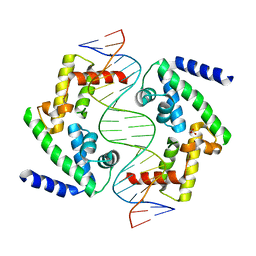 | | DNA binding domain of HNF-1A bound to P2-HNF4A promoter DNA variant (P2 -181G>T) | | Descriptor: | Chains: E, Chains: F, GLYCEROL, ... | | Authors: | Kind, L, Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular mechanism of HNF-1A-mediated HNF4A gene regulation and promoter-driven HNF4A-MODY diabetes.
JCI Insight, 9, 2024
|
|
8PI7
 
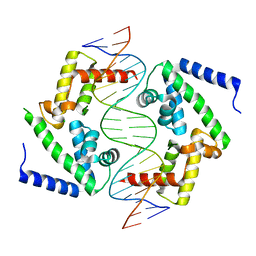 | | DNA binding domain of HNF-1A bound to P2-HNF4A promoter DNA variant (P2 -169C>T) | | Descriptor: | CHLORIDE ION, Chains: E, Chains: F, ... | | Authors: | Kind, L, Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular mechanism of HNF-1A-mediated HNF4A gene regulation and promoter-driven HNF4A-MODY diabetes.
JCI Insight, 9, 2024
|
|
8PI9
 
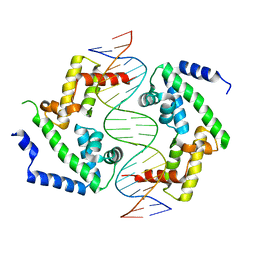 | | DNA binding domain of HNF-1A bound to P2-HNF4A promoter DNA variant (P2 -181G>A) | | Descriptor: | Chains: E, Chains: F, Hepatocyte nuclear factor 1-alpha | | Authors: | Kind, L, Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular mechanism of HNF-1A-mediated HNF4A gene regulation and promoter-driven HNF4A-MODY diabetes.
JCI Insight, 9, 2024
|
|
8PI8
 
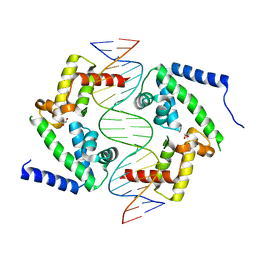 | | DNA binding domain of HNF-1A bound to P2-HNF4A promoter DNA | | Descriptor: | Chains: E, Chains: F, GLYCEROL, ... | | Authors: | Kind, L, Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism of HNF-1A-mediated HNF4A gene regulation and promoter-driven HNF4A-MODY diabetes.
JCI Insight, 9, 2024
|
|
6TA2
 
 | | Human NAMPT in complex with nicotinic acid mononucleotide and phosphate | | Descriptor: | CHLORIDE ION, GLYCEROL, NICOTINATE MONONUCLEOTIDE, ... | | Authors: | Houry, D, Raasakka, A, Kursula, P, Ziegler, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification of structural determinants of NAMPT activity and substrate selectivity
To Be Published
|
|
6TAC
 
 | | Human NAMPT deletion mutant in complex with nicotinamide mononucleotide, pyrophosphate, and Mg2+ | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Houry, D, Raasakka, A, Kursula, P, Ziegler, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of structural determinants of NAMPT activity and substrate selectivity
To Be Published
|
|
4WC9
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation F235L | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
