1V81
 
 | |
1UL5
 
 | | Solution structure of the DNA-binding domain of squamosa promoter binding protein-like 7 | | Descriptor: | ZINC ION, squamosa promoter binding protein-like 7 | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors.
J.Mol.Biol., 337, 2004
|
|
2ZDJ
 
 | | Crystal Structure of TTMA177, a Hypothetical Protein from Thermus thermophilus phage TMA | | Descriptor: | hypothetical protein TTMA177 | | Authors: | Agari, Y, Tamakoshi, M, Yamagishi, A, Shinkai, A, Ebihara, A, Yokoyama, S, Kuramitsu, S, Oshima, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-11-26 | | Release date: | 2008-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of TTMA177, a Hypothetical Protein from Thermus thermophilus phage TMA
To be Published
|
|
2ZOP
 
 | | X-ray crystal structure of a CRISPR-associated Cmr5 family protein from Thermus thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHB164, SULFATE ION | | Authors: | Sakamoto, K, Agari, Y, Shinkai, A, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-05-28 | | Release date: | 2009-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of a CRISPR-associated RAMP superfamily protein, Cmr5, from Thermus thermophilus HB8
Proteins, 75, 2009
|
|
2YRK
 
 | | Solution structure of the zf-C2H2 domain in zinc finger homeodomain 4 | | Descriptor: | ZINC ION, Zinc finger homeobox protein 4 | | Authors: | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the zf-C2H2 domain in zinc finger homeodomain 4
To be Published
|
|
1VBN
 
 | | Escherichia coli tyrosyl-tRNA synthetase mutant complexed with Tyr-AMS | | Descriptor: | 5'-O-[N-(L-TYROSYL)SULFAMOYL]ADENOSINE, Tyrosyl-tRNA synthetase | | Authors: | Kobayashi, T, Sakamoto, K, Takimura, T, Kamata, K, Sekine, R, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-27 | | Release date: | 2005-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1VC4
 
 | | Crystal Structure of Indole-3-Glycerol Phosphate Synthase (TrpC) from Thermus Thermophilus At 1.8 A Resolution | | Descriptor: | ACETIC ACID, GLYCEROL, Indole-3-Glycerol Phosphate Synthase, ... | | Authors: | Bagautdinov, B, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-04 | | Release date: | 2004-03-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of indole-3-glycerol phosphate synthase from Thermus thermophilus HB8: implications for thermal stability.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2YRB
 
 | |
2YS5
 
 | | Solution structure of the complex of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of hALK | | Descriptor: | ALK tyrosine kinase receptor, Fibroblast growth factor receptor substrate 3 | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
2YT2
 
 | | Solution structure of the chimera of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of hALK | | Descriptor: | Fibroblast growth factor receptor substrate 3 and ALK tyrosine kinase receptor | | Authors: | Li, H, Koshiba, S, Tomizawa, T, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
2YTY
 
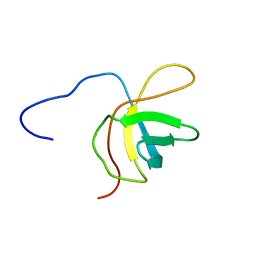 | | Solution structure of the fourth cold-shock domain of the human KIAA0885 protein (UNR protein) | | Descriptor: | Cold shock domain-containing protein E1 | | Authors: | Goroncy, A.K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
2YWO
 
 | |
1V80
 
 | |
2YVO
 
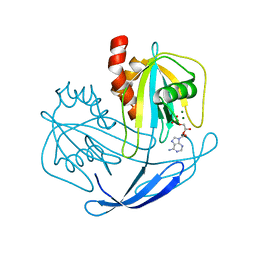 | | Crystal structure of NDX2 in complex with MG2+ and AMP from thermus thermophilus HB8 | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, MutT/nudix family protein | | Authors: | Wakamatsu, T, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-13 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural basis for different substrate specificities of two ADP-ribose pyrophosphatases from Thermus thermophilus HB8
J.Bacteriol., 190, 2008
|
|
1V3R
 
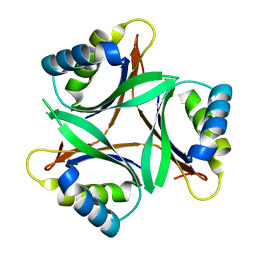 | | Crystal structure of TT1020 from Thermus thermophilus HB8 | | Descriptor: | Nitrogen regulatory protein P-II | | Authors: | Wang, H, Sakai, H, Takemoto-Hori, C, Kaminishi, T, Yamaguchi, H, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-05 | | Release date: | 2004-11-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the signal transducing protein GlnK from Thermus thermophilus HB8.
J.Struct.Biol., 149, 2005
|
|
2ZMU
 
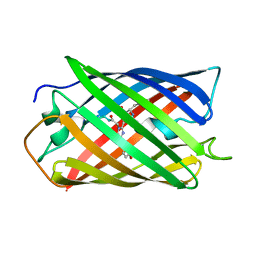 | | Crystal Structure of Monomeric Kusabira-Orange (MKO), Orange-Emitting GFP-like Protein, at pH 9.1 | | Descriptor: | Fluorescent protein | | Authors: | Kikuchi, A, Fukumura, E, Karasawa, S, Mizuno, H, Miyawaki, A, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-21 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterization of a Thiazoline-Containing Chromophore in an Orange Fluorescent Protein, Monomeric Kusabira Orange
Biochemistry, 47, 2008
|
|
1V5W
 
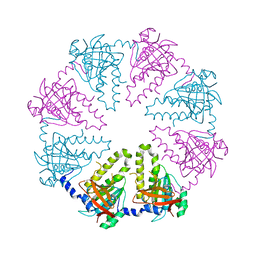 | | Crystal structure of the human Dmc1 protein | | Descriptor: | Meiotic recombination protein DMC1/LIM15 homolog | | Authors: | Kinebuchi, T, Kagawa, W, Enomoto, R, Ikawa, S, Shibata, T, Kurumizaka, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-26 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein dmc1
Mol.Cell, 14, 2004
|
|
1V2Z
 
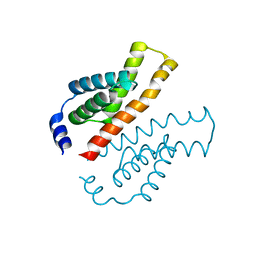 | | Crystal structure of the C-terminal domain of Thermosynechococcus elongatus BP-1 KaiA | | Descriptor: | circadian clock protein KaiA homolog | | Authors: | Uzumaki, T, Fujita, M, Nakatsu, T, Hayashi, F, Shibata, H, Itoh, N, Kato, H, Ishiura, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-20 | | Release date: | 2004-06-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the C-terminal clock-oscillator domain of the cyanobacterial KaiA protein
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
1V6T
 
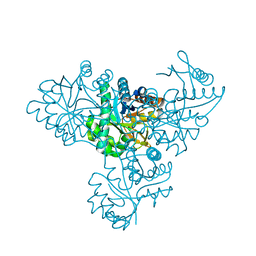 | |
1V9M
 
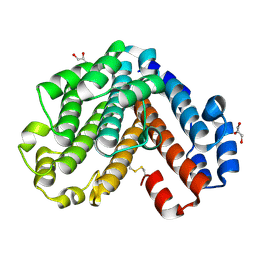 | | Crystal structure of the C subunit of V-type ATPase from Thermus thermophilus | | Descriptor: | GLYCEROL, V-type ATP synthase subunit C | | Authors: | Numoto, N, Kita, A, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the C subunit of V-type ATPase from Thermus thermophilus at 1.85 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1VBH
 
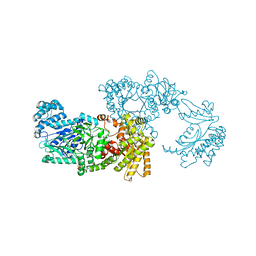 | | Pyruvate Phosphate Dikinase with bound Mg-PEP from Maize | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE, SULFATE ION, ... | | Authors: | Nakanishi, T, Nakatsu, T, Matsuoka, M, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of pyruvate phosphate dikinase from maize revealed an alternative conformation in the swiveling-domain motion
Biochemistry, 44, 2005
|
|
1UL4
 
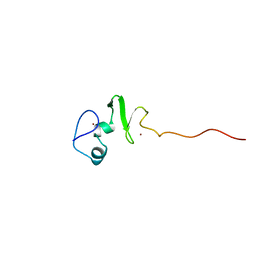 | | Solution structure of the DNA-binding domain of squamosa promoter binding protein-like 4 | | Descriptor: | ZINC ION, squamosa promoter binding protein-like 4 | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors.
J.Mol.Biol., 337, 2004
|
|
1V2B
 
 | | Crystal Structure of PsbP Protein in the Oxygen-Evolving Complex of Photosystem II from Higher Plants | | Descriptor: | 23-kDa polypeptide of photosystem II oxygen-evolving complex, SULFATE ION, alpha-D-glucopyranose | | Authors: | Ifuku, K, Nakatsu, T, Kato, H, Sato, F, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-14 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the PsbP protein of photosystem II from Nicotiana tabacum
Embo Rep., 5, 2004
|
|
1UMH
 
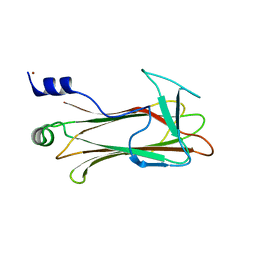 | | Structural basis of sugar-recognizing ubiquitin ligase | | Descriptor: | F-box only protein 2, NICKEL (II) ION | | Authors: | Mizushima, T, Hirao, T, Yoshida, Y, Lee, S.J, Chiba, T, Iwai, K, Yamaguchi, Y, Kato, K, Tsukihara, T, Tanaka, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-01 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of sugar-recognizing ubiquitin ligase
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
3AOH
 
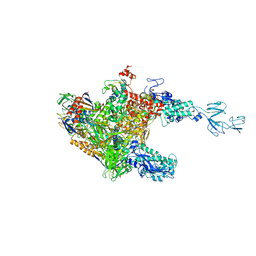 | | RNA polymerase-Gfh1 complex (Crystal type 1) | | Descriptor: | Anti-cleavage anti-GreA transcription factor Gfh1, DNA (5'-D(*GP*GP*TP*CP*TP*GP*TP*AP*TP*CP*AP*CP*GP*AP*GP*CP*CP*AP*CP*CP*GP*CP*CP*GP*CP*AP*T)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Tagami, S, Sekine, S, Kumarevel, T, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-28 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein
Nature, 468, 2010
|
|
