2LA0
 
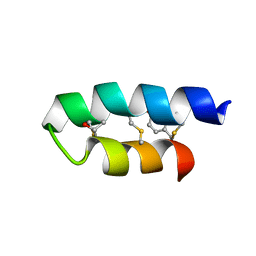 | | Trn- peptide of the two-component bacteriocin Thuricin CD | | Descriptor: | Uncharacterized protein | | Authors: | Sit, C.S, Mckay, R.T, Hill, C, Ross, R.P, Vederas, J.C. | | Deposit date: | 2011-02-27 | | Release date: | 2012-01-11 | | Last modified: | 2018-08-22 | | Method: | SOLUTION NMR | | Cite: | The 3D structure of thuricin CD, a two-component bacteriocin with cysteine sulfur to alpha-carbon cross-links.
J.Am.Chem.Soc., 133, 2011
|
|
2LHR
 
 | |
4FEN
 
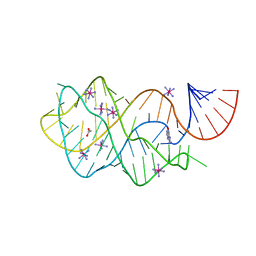 | | Crystal structure of the A24U/U25A/A46G mutant xpt-pbuX guanine riboswitch aptamer domain in complex with hypoxanthine | | Descriptor: | A24U/U25A/A46G mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
4FRN
 
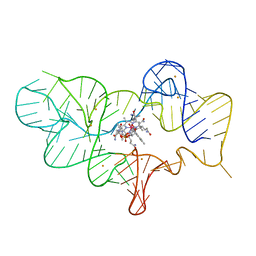 | | Crystal structure of the cobalamin riboswitch regulatory element | | Descriptor: | BARIUM ION, Cobalamin riboswitch aptamer domain, Hydroxocobalamin | | Authors: | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | Deposit date: | 2012-06-26 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
4FEJ
 
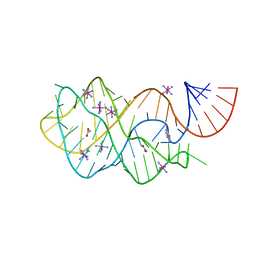 | | Crystal structure of the A24U mutant xpt-pbuX guanine riboswitch aptamer domain in complex with hypoxanthine | | Descriptor: | A24U mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
2IEF
 
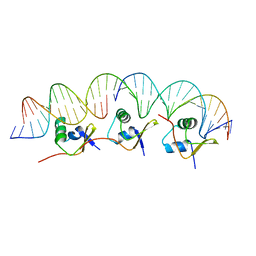 | | Structure of the cooperative Excisionase (Xis)-DNA complex reveals a micronucleoprotein filament | | Descriptor: | 15-mer DNA, 19-mer DNA, 34-mer DNA, ... | | Authors: | Abbani, M.A, Papagiannis, C.V, Sam, M.D, Cascio, D, Johnson, R.C, Clubb, R.T. | | Deposit date: | 2006-09-18 | | Release date: | 2007-02-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structure of the cooperative Xis-DNA complex reveals a micronucleoprotein filament that regulates phage lambda intasome assembly.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4FEP
 
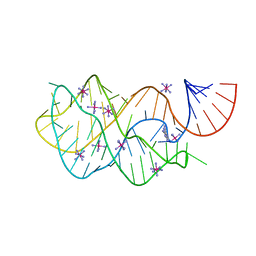 | | Crystal structure of the A24U/U25A/A46G/C74U mutant xpt-pbuX guanine riboswitch aptamer domain in complex with 2,6-diaminopurine | | Descriptor: | 9H-PURINE-2,6-DIAMINE, A24U/U25A/A46G/C74U mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain, COBALT HEXAMMINE(III) | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
4FE5
 
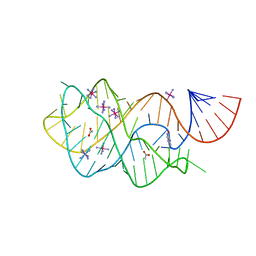 | | Crystal structure of the xpt-pbuX guanine riboswitch aptamer domain in complex with hypoxanthine | | Descriptor: | ACETATE ION, COBALT HEXAMMINE(III), HYPOXANTHINE, ... | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-29 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine.
Nature, 432, 2004
|
|
4FEO
 
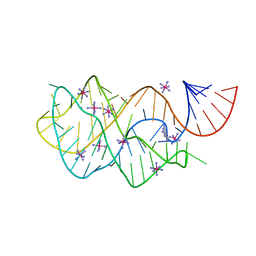 | | Crystal structure of the AU25A/A46G/C74U mutant xpt-pbuX guanine riboswitch aptamer domain in complex with 2,6-diaminopurine | | Descriptor: | 9H-PURINE-2,6-DIAMINE, COBALT HEXAMMINE(III), U25A/A46G/C74U mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
2M60
 
 | | Enterocin 7B | | Descriptor: | Enterocin JSB | | Authors: | Lohans, C.T, Towle, K.M, Miskolzie, M, McKay, R.T, van Belkum, M.J, McMullen, L.M, Vederas, J.C. | | Deposit date: | 2013-03-18 | | Release date: | 2013-06-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the Linear Leaderless Bacteriocins Enterocin 7A and 7B Resemble Carnocyclin A, a Circular Antimicrobial Peptide
Biochemistry, 52, 2013
|
|
2JMF
 
 | | Solution structure of the Su(dx) WW4- Notch PY peptide complex | | Descriptor: | E3 ubiquitin-protein ligase suppressor of deltex, Neurogenic locus Notch protein | | Authors: | Avis, J.M, Blankley, R.T, Jennings, M.D, Golovanov, A.P. | | Deposit date: | 2006-11-05 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and Autoregulation of Notch Binding by Tandem WW Domains in Suppressor of Deltex
J.Biol.Chem., 282, 2007
|
|
4GMA
 
 | | Crystal structure of the adenosylcobalamin riboswitch | | Descriptor: | Adenosylcobalamin, Adenosylcobalamin riboswitch | | Authors: | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | Deposit date: | 2012-08-15 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
2M5Z
 
 | | Enterocin 7A | | Descriptor: | Enterocin JSA | | Authors: | Lohans, C.T, Towle, K.M, Miskolzie, M, McKay, R.T, van Belkum, M.J, McMullen, L.M, Vederas, J.C. | | Deposit date: | 2013-03-16 | | Release date: | 2013-06-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the Linear Leaderless Bacteriocins Enterocin 7A and 7B Resemble Carnocyclin A, a Circular Antimicrobial Peptide
Biochemistry, 52, 2013
|
|
4E76
 
 | |
4E78
 
 | |
2LBZ
 
 | | Thurincin H | | Descriptor: | Thuricin17 | | Authors: | Sit, C.S, van Belkum, M.J, Mckay, R.T, Worobo, R.W, Vederas, J.C. | | Deposit date: | 2011-04-10 | | Release date: | 2012-01-18 | | Last modified: | 2018-08-22 | | Method: | SOLUTION NMR | | Cite: | The 3D solution structure of thurincin H, a bacteriocin with four sulfur to alpha-carbon crosslinks.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2LN7
 
 | |
2L9X
 
 | | Trn- peptide of the two-component bacteriocin Thuricin CD | | Descriptor: | Uncharacterized protein | | Authors: | Sit, C.S, Mckay, R.T, Hill, C, Ross, R.P, Vederas, J.C. | | Deposit date: | 2011-02-25 | | Release date: | 2012-01-11 | | Last modified: | 2018-08-22 | | Method: | SOLUTION NMR | | Cite: | The 3D structure of thuricin CD, a two-component bacteriocin with cysteine sulfur to alpha-carbon cross-links.
J.Am.Chem.Soc., 133, 2011
|
|
2MP2
 
 | | Solution structure of SUMO dimer in complex with SIM2-3 from RNF4 | | Descriptor: | E3 ubiquitin-protein ligase RNF4, Small ubiquitin-related modifier 3 | | Authors: | Xu, Y, Plechanovov, A, Simpson, P, Marchant, J, Leidecker, O, Sebastian, K, Hay, R.T, Matthews, S.J. | | Deposit date: | 2014-05-09 | | Release date: | 2014-07-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into SUMO chain recognition and manipulation by the ubiquitin ligase RNF4.
Nat Commun, 5, 2014
|
|
2NPF
 
 | | Structure of eEF2 in complex with moriniafungin | | Descriptor: | (1S,4R,5R,9S,11S)-2-({[(2S,5R,6R,7R,9S,10R)-2-(7-CARBOXYHEPTYL)-6-HYDROXY-10-METHOXY-9-METHYL-3-OXO-1,4,8-TRIOXASPIRO[4 .5]DEC-7-YL]OXY}METHYL)-9-FORMYL-13-ISOPROPYL-5-METHYLTETRACYCLO[7.4.0.02,11.04.8]TRIDEC-12-ENE-1-CARBOXYLIC ACID, Elongation factor 2, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Soe, R, Mosley, R.T, Andersen, G.R. | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Sordarin derivatives induce a novel conformation of the yeast ribosome translocation factor eEF2
J.Biol.Chem., 282, 2007
|
|
4ERJ
 
 | |
4FRG
 
 | | Crystal structure of the cobalamin riboswitch aptamer domain | | Descriptor: | Hydroxocobalamin, IRIDIUM (III) ION, MAGNESIUM ION, ... | | Authors: | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | Deposit date: | 2012-06-26 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
4G34
 
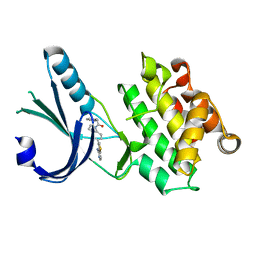 | | Crystal Structure of GSK6924 Bound to PERK (R587-R1092, delete A660-T867) at 2.70 A Resolution | | Descriptor: | 1-[5-(4-aminothieno[3,2-c]pyridin-3-yl)-2,3-dihydro-1H-indol-1-yl]-2-phenylethanone, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Gampe, R.T, Axten, J.M. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 55, 2012
|
|
4G31
 
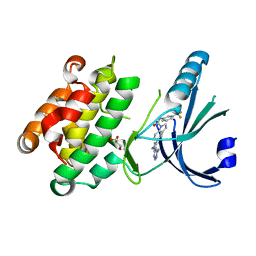 | | Crystal Structure of GSK6414 Bound to PERK (R587-R1092, delete A660-T867) at 2.28 A Resolution | | Descriptor: | 1-[5-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-2,3-dihydro-1H-indol-1-yl]-2-[3-(trifluoromethyl)phenyl]ethanone, Eukaryotic translation initiation factor 2-alpha kinase 3, GLYCEROL | | Authors: | Gampe, R.T, Axten, J.M. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 55, 2012
|
|
4I5O
 
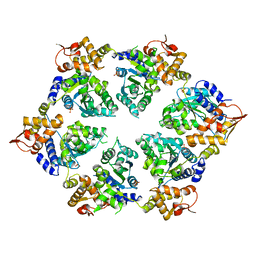 | | Crystal Structure of W-W-R ClpX Hexamer | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.4787 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
