6A8X
 
 | | Crystal structure of a apo form cyclase from Fischerella sp. | | Descriptor: | CALCIUM ION, aromatic prenyltransferase | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-07-11 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5ZZD
 
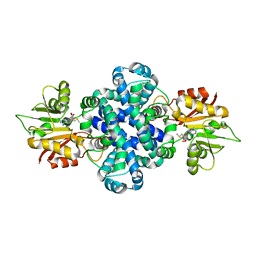 | | Crystal structure of a protein from Aspergillus flavus | | Descriptor: | O-methyltransferase lepI, S-ADENOSYLMETHIONINE | | Authors: | Chang, Z.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-05-31 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of LepI, a multifunctional SAM-dependent enzyme which catalyzes pericyclic reactions in leporin biosynthesis.
Org.Biomol.Chem., 17, 2019
|
|
5ZXA
 
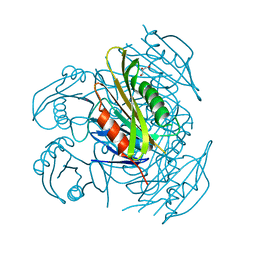 | | Crystal structure of fibronectin-binding protein Apa mutant from Mycobacterium tuberculosis | | Descriptor: | Alanine and proline-rich secreted protein Apa, GLYCEROL, MERCURY (II) ION | | Authors: | Gao, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-05-18 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Functional and structural investigations of fibronectin-binding protein Apa from Mycobacterium tuberculosis.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
5ZZJ
 
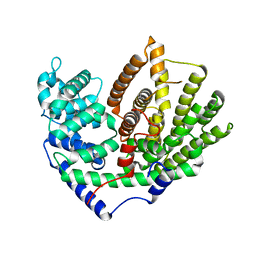 | |
6A2A
 
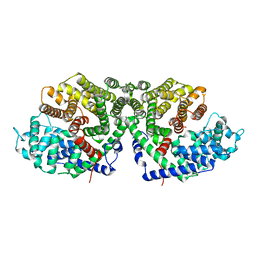 | | Crystal structure of a synthase 2 from santalum album | | Descriptor: | Sesquisabinene B synthase 2 | | Authors: | Han, X, Ko, T.P, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-06-09 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of a synthase 2 from santalum album
to be published
|
|
6A1D
 
 | | Crystal structure of a synthase 1 from Santalum album in complex with ligand | | Descriptor: | MAGNESIUM ION, SULFATE ION, Sesquisabinene B synthase 1, ... | | Authors: | Han, X, Ko, T.P, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-06-07 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of a synthase 1 from Santalum album in complex with ligand
To Be Published
|
|
5ZX9
 
 | | Crystal structure of apo form fibronectin-binding protein Apa from Mycobacterium tuberculosis | | Descriptor: | Alanine and proline-rich secreted protein Apa, GLYCEROL | | Authors: | Gao, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-05-18 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Functional and structural investigations of fibronectin-binding protein Apa from Mycobacterium tuberculosis.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
5ZXK
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 3 member A1 | | Authors: | Yang, Y.Y, Liu, W.D, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
6A1E
 
 | | Crystal structure of a synthase 1 from Santalum album in complex with ligand fps | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CARBONATE ION, GLYCEROL, ... | | Authors: | Han, X, Ko, T.P, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-06-07 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of a synthase 1 from Santalum album in complex with ligand
To Be Published
|
|
6A99
 
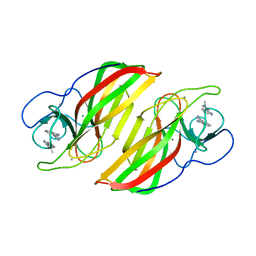 | | Crystal structure of a Stig cyclases Fisc from Fischerella sp. TAU in complex with (3Z)-3-(1-methyl-2-pyrrolidinylidene)-3H-indole | | Descriptor: | (3~{Z})-3-(1-methylpyrrolidin-2-ylidene)indole, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-07-12 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6ADU
 
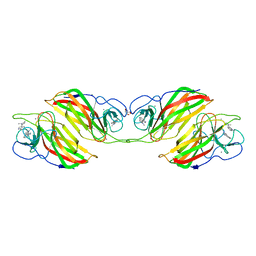 | | Crystal structure of an enzyme in complex with ligand C | | Descriptor: | (3~{Z})-3-(1-methylpyrrolidin-2-ylidene)indole, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Tan, X.K, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-08-02 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement.
Angew.Chem.Int.Ed.Engl., 57, 2018
|
|
6A3X
 
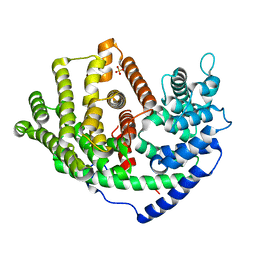 | | Crystal structure of a synthase 1 from Santalum album in complex with ligand s | | Descriptor: | SULFATE ION, Sabinene, Sesquisabinene B synthase 1 | | Authors: | Han, X, Ko, T.P, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-06-18 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a synthase 1 from Santalum album in complex with ligand s
To Be Published
|
|
6A9F
 
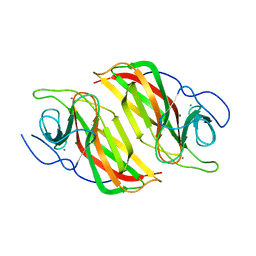 | | Crystal structure of a cyclase from Fischerella sp. TAU in complex with 4-(1H-Indol-3-yl)butan-2-one | | Descriptor: | 4-(1~{H}-indol-3-yl)butan-2-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-07-13 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
1VWA
 
 | | STREPTAVIDIN-FSHPQNT | | Descriptor: | PEPTIDE LIGAND CONTAINING HPQ, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In crystals of complexes of streptavidin with peptide ligands containing the HPQ sequence the pKa of the peptide histidine is less than 3.0.
J.Biol.Chem., 272, 1997
|
|
1VWH
 
 | |
1VWE
 
 | | STREPTAVIDIN-CYCLO-AC-[CHPQFC]-NH2, PH 3.6 | | Descriptor: | PEPTIDE LIGAND CONTAINING HPQ, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | In crystals of complexes of streptavidin with peptide ligands containing the HPQ sequence the pKa of the peptide histidine is less than 3.0.
J.Biol.Chem., 272, 1997
|
|
1VWM
 
 | | STREPTAVIDIN-CYCLO-AC-[CHPQFC]-NH2, PH 4.2 | | Descriptor: | PEPTIDE LIGAND CONTAINING HPQ, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In crystals of complexes of streptavidin with peptide ligands containing the HPQ sequence the pKa of the peptide histidine is less than 3.0.
J.Biol.Chem., 272, 1997
|
|
1VWL
 
 | |
1VWK
 
 | | STREPTAVIDIN-CYCLO-[5-S-VALERAMIDE-HPQGPPC]K-NH2 | | Descriptor: | PENTANOIC ACID, PEPTIDE LIGAND CONTAINING HPQ, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | In crystals of complexes of streptavidin with peptide ligands containing the HPQ sequence the pKa of the peptide histidine is less than 3.0.
J.Biol.Chem., 272, 1997
|
|
1VWQ
 
 | |
1VWO
 
 | |
1VWI
 
 | |
5Z5N
 
 | | Crystal structure of ConA-R1M | | Descriptor: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | Authors: | Chen, G.S, Gan, J.H, Hu, R.T. | | Deposit date: | 2018-01-18 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of ConA-R1M (Only Mannose)
To Be Published
|
|
5Z97
 
 | | Crystal structure of a lactonase double mutant in complex with ligand N | | Descriptor: | (3S,11E)-14,16-dihydroxy-3-methyl-3,4,5,6,9,10-hexahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Lactonase for protein, SULFATE ION | | Authors: | Zheng, Y.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-02-02 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
5Z5P
 
 | | Crystal structure of ConA-R3M | | Descriptor: | 2-[2-(2-{4-[(alpha-D-mannopyranosyloxy)methyl]-1H-1,2,3-triazol-1-yl}ethoxy)ethoxy]ethyl 2-[3,6-bis(diethylamino)-9H-xanthen-9-yl]benzoate, CALCIUM ION, Concanavalin-A, ... | | Authors: | Chen, G.S, Gan, J.H, Hu, R.T. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of ConA-R3M
To Be Published
|
|
