1IRL
 
 | | THE SOLUTION STRUCTURE OF THE F42A MUTANT OF HUMAN INTERLEUKIN 2 | | 分子名称: | INTERLEUKIN-2 | | 著者 | Mott, H.R, Baines, B.S, Hall, R.M, Cooke, R.M, Driscoll, P.C, Weir, M.P, Campbell, I.D. | | 登録日 | 1995-08-25 | | 公開日 | 1995-12-07 | | 最終更新日 | 2021-11-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the F42A mutant of human interleukin 2.
J.Mol.Biol., 247, 1995
|
|
4ICD
 
 | | REGULATION OF ISOCITRATE DEHYDROGENASE BY PHOSPHORYLATION INVOLVES NO LONG-RANGE CONFORMATIONAL CHANGE IN THE FREE ENZYME | | 分子名称: | PHOSPHORYLATED ISOCITRATE DEHYDROGENASE | | 著者 | Hurley, J.H, Dean, A.M, Thorsness, P.E, Koshlandjunior, D.E, Stroud, R.M. | | 登録日 | 1989-12-28 | | 公開日 | 1991-01-15 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Regulation of isocitrate dehydrogenase by phosphorylation involves no long-range conformational change in the free enzyme.
J.Biol.Chem., 265, 1990
|
|
1TGN
 
 | |
1DTT
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 (PETT130A94) | | 分子名称: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | 著者 | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | 登録日 | 2000-01-13 | | 公開日 | 2000-04-02 | | 最終更新日 | 2014-11-12 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
1DTQ
 
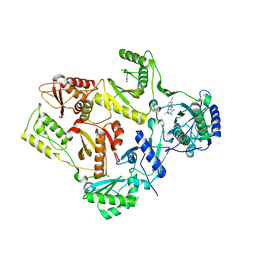 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-1 (PETT131A94) | | 分子名称: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-NITRILOMETHYL-PYRIDYL]-THIOUREA | | 著者 | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | 登録日 | 2000-01-13 | | 公開日 | 2000-03-20 | | 最終更新日 | 2014-11-12 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
1XEE
 
 | | Solution structure of the Chemotaxis Inhibitory Protein of Staphylococcus aureus | | 分子名称: | chemotaxis-inhibiting protein CHIPS | | 著者 | Haas, P.J, de Haas, C.J, Poppelier, M.J, van Kessel, K.P, van Strijp, J.A, Dijkstra, K, Scheek, R.M, Fan, H, Kruijtzer, J.A, Liskamp, R.M, Kemmink, J. | | 登録日 | 2004-09-10 | | 公開日 | 2005-09-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of the C5a receptor-blocking domain of chemotaxis inhibitory protein of Staphylococcus aureus is related to a group of immune evasive molecules
J.Mol.Biol., 353, 2005
|
|
4K1C
 
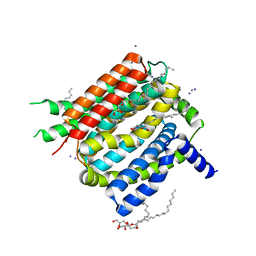 | | VCX1 Calcium/Proton Exchanger | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, IODIDE ION, ... | | 著者 | Waight, A.B, Pedersen, B.P, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | 登録日 | 2013-04-04 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for alternating access of a eukaryotic calcium/proton exchanger.
Nature, 499, 2013
|
|
4K2D
 
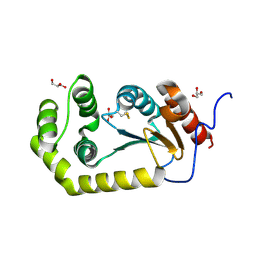 | | Crystal structure of Burkholderia Pseudomallei DsbA | | 分子名称: | GLYCEROL, Thiol:disulfide interchange protein | | 著者 | McMahon, R.M. | | 登録日 | 2013-04-09 | | 公開日 | 2013-08-14 | | 最終更新日 | 2014-02-12 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Disarming Burkholderia pseudomallei: Structural and Functional Characterization of a Disulfide Oxidoreductase (DsbA) Required for Virulence In Vivo.
Antioxid Redox Signal, 20, 2014
|
|
4K0E
 
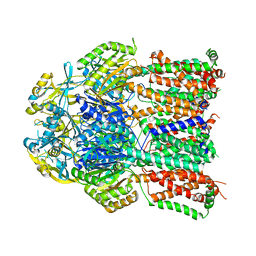 | | X-ray crystal structure of a heavy metal efflux pump, crystal form II | | 分子名称: | Heavy metal cation tricomponent efflux pump ZneA(CzcA-like), ZINC ION | | 著者 | Pak, J.E, Ngonlong Ekende, E, Vandenbussche, G, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | 登録日 | 2013-04-03 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.713 Å) | | 主引用文献 | Structures of intermediate transport states of ZneA, a Zn(II)/proton antiporter.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K0J
 
 | | X-ray crystal structure of a heavy metal efflux pump, crystal form I | | 分子名称: | Heavy metal cation tricomponent efflux pump ZneA(CzcA-like), ZINC ION | | 著者 | Pak, J.E, Stroud, R.M, Ngonlong Ekende, E, Vandenbussche, G, Center for Structures of Membrane Proteins (CSMP) | | 登録日 | 2013-04-04 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures of intermediate transport states of ZneA, a Zn(II)/proton antiporter.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5W7P
 
 | | Crystal structure of OxaC | | 分子名称: | OxaC, S-ADENOSYLMETHIONINE | | 著者 | Newmister, S.A, Romminger, S, Schmidt, J.J, Williams, R.M, Smith, J.L, Berlinck, R.G.S, Sherman, D.H. | | 登録日 | 2017-06-20 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Unveiling sequential late-stage methyltransferase reactions in the meleagrin/oxaline biosynthetic pathway.
Org. Biomol. Chem., 16, 2018
|
|
7MKE
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to lambda PR promoter DNA (class 2) | | 分子名称: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Saecker, R.M, Darst, S.A, Chen, J. | | 登録日 | 2021-04-23 | | 公開日 | 2021-09-29 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MKJ
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to T7A1 promoter DNA | | 分子名称: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Saecker, R.M, Darst, S.A, Chen, J. | | 登録日 | 2021-04-23 | | 公開日 | 2021-09-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MMM
 
 | | LyeTx I | | 分子名称: | Toxin LyeTx 1 | | 著者 | Verly, R.M. | | 登録日 | 2021-04-29 | | 公開日 | 2021-09-29 | | 最終更新日 | 2021-11-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | LyeTx I, a potent antimicrobial peptide from the venom of the spider Lycosa erythrognatha
Amino Acids, 39, 2010
|
|
7MKI
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to lambda PR (-5G to C) promoter DNA | | 分子名称: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Saecker, R.M, Darst, S.A, Chen, J. | | 登録日 | 2021-04-23 | | 公開日 | 2021-09-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MKD
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to lambda PR promoter DNA (class 1) | | 分子名称: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Saecker, R.M, Darst, S.A, Chen, J. | | 登録日 | 2021-04-23 | | 公開日 | 2021-09-29 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5WN9
 
 | | Structure of antibody 2D10 bound to the central conserved region of RSV G | | 分子名称: | Major surface glycoprotein G, scFv 2D10 | | 著者 | Fedechkin, S.O, George, N.L, Wolff, J.T, Kauvar, L.M, DuBois, R.M. | | 登録日 | 2017-07-31 | | 公開日 | 2018-03-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.551 Å) | | 主引用文献 | Structures of respiratory syncytial virus G antigen bound to broadly neutralizing antibodies.
Sci Immunol, 3, 2018
|
|
7CCR
 
 | | Structure of the 2:2 cGAS-nucleosome complex | | 分子名称: | Cyclic GMP-AMP synthase, DNA (147-MER), Histone H2A type 1-B/E, ... | | 著者 | Cao, D, Han, X, Fan, X, Xu, R.M, Zhang, X. | | 登録日 | 2020-06-17 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Structural basis for nucleosome-mediated inhibition of cGAS activity.
Cell Res., 30, 2020
|
|
5W7K
 
 | | Crystal structure of OxaG | | 分子名称: | CHLORIDE ION, OxaG, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Newmister, S.A, Romminger, S, Schmidt, J.J, Williams, R.M, Smith, J.L, Berlinck, R.G.S, Sherman, D.H. | | 登録日 | 2017-06-20 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.994 Å) | | 主引用文献 | Unveiling sequential late-stage methyltransferase reactions in the meleagrin/oxaline biosynthetic pathway.
Org. Biomol. Chem., 16, 2018
|
|
5W7S
 
 | | Crystal structure of OxaC in complex with sinefungin and meleagrin | | 分子名称: | (3E,7aR,12aS)-6-hydroxy-3-[(1H-imidazol-4-yl)methylidene]-12-methoxy-7a-(2-methylbut-3-en-2-yl)-7a,12-dihydro-1H,5H-imidazo[1',2':1,2]pyrido[2,3-b]indole-2,5(3H)-dione, OxaC, SINEFUNGIN | | 著者 | Newmister, S.A, Romminger, S, Schmidt, J.J, Williams, R.M, Smith, J.L, Berlinck, R.G.S, Sherman, D.H. | | 登録日 | 2017-06-20 | | 公開日 | 2018-06-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.948 Å) | | 主引用文献 | Unveiling sequential late-stage methyltransferase reactions in the meleagrin/oxaline biosynthetic pathway.
Org. Biomol. Chem., 16, 2018
|
|
7CCQ
 
 | | Structure of the 1:1 cGAS-nucleosome complex | | 分子名称: | Cyclic GMP-AMP synthase, DNA (147-MER), Histone H2A type 1-B/E, ... | | 著者 | Cao, D, Han, X, Fan, X, Xu, R.M, Zhang, X. | | 登録日 | 2020-06-17 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for nucleosome-mediated inhibition of cGAS activity.
Cell Res., 30, 2020
|
|
4K5Y
 
 | | Crystal structure of human corticotropin-releasing factor receptor 1 (CRF1R) in complex with the antagonist CP-376395 | | 分子名称: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6-dimethyl-N-(pentan-3-yl)-2-(2,4,6-trimethylphenoxy)pyridin-4-amine, ... | | 著者 | Hollenstein, K, Kean, J, Bortolato, A, Cheng, R.K.Y, Dore, A.S, Jazayeri, A, Cooke, R.M, Weir, M, Marshall, F.H. | | 登録日 | 2013-04-15 | | 公開日 | 2013-07-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.977 Å) | | 主引用文献 | Structure of class B GPCR corticotropin-releasing factor receptor 1.
Nature, 499, 2013
|
|
1F09
 
 | | CRYSTAL STRUCTURE OF THE GREEN FLUORESCENT PROTEIN (GFP) VARIANT YFP-H148Q WITH TWO BOUND IODIDES | | 分子名称: | GREEN FLUORESCENT PROTEIN, IODIDE ION | | 著者 | Wachter, R.M, Yarbrough, D, Kallio, K, Remington, S.J. | | 登録日 | 2000-05-15 | | 公開日 | 2000-11-17 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Crystallographic and energetic analysis of binding of selected anions to the yellow variants of green fluorescent protein.
J.Mol.Biol., 301, 2000
|
|
5W7M
 
 | | Crystal structure of RoqN | | 分子名称: | Glandicoline B O-methyltransferase roqN, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Newmister, S.A, Romminger, S, Schmidt, J.J, Williams, R.M, Smith, J.L, Berlinck, R.G.S, Sherman, D.H. | | 登録日 | 2017-06-20 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Unveiling sequential late-stage methyltransferase reactions in the meleagrin/oxaline biosynthetic pathway.
Org. Biomol. Chem., 16, 2018
|
|
1F0B
 
 | |
