5N7Y
 
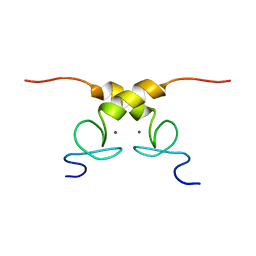 | | Solution structure of B. subtilis Sigma G inhibitor CsfB | | Descriptor: | Anti-sigma-G factor Gin, ZINC ION | | Authors: | Martinez-Lumbreras, S, Alfano, C, Atkinson, A, Isaacson, R.L. | | Deposit date: | 2017-02-21 | | Release date: | 2018-02-28 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Insights into Bacillus subtilis Sigma Factor Inhibitor, CsfB.
Structure, 26, 2018
|
|
5NP0
 
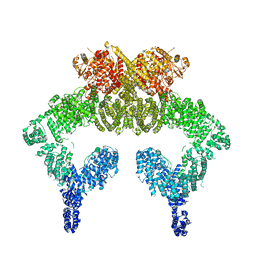 | | Closed dimer of human ATM (Ataxia telangiectasia mutated) | | Descriptor: | Serine-protein kinase ATM | | Authors: | Baretic, D, Pollard, H.K, Fisher, D.I, Johnson, C.M, Santhanam, B, Truman, C.M, Kouba, T, Fersht, A.R, Phillips, C, Williams, R.L. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of closed and open conformations of dimeric human ATM.
Sci Adv, 3, 2017
|
|
5O8A
 
 | | Crystal Structure of rsEGFP2 in the non-fluorescent off-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
5NWT
 
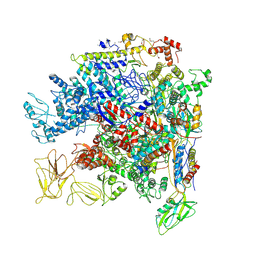 | | Crystal Structure of Escherichia coli RNA polymerase - Sigma54 Holoenzyme complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhang, X, Buck, M, Darbari, V.C, Yang, Y, Zhang, N, Lu, D, Glyde, R, Wang, Y, Winkelman, J, Gourse, R.L, Murakami, K.S. | | Deposit date: | 2017-05-08 | | Release date: | 2017-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.76 Å) | | Cite: | TRANSCRIPTION. Structures of the RNA polymerase-Sigma54 reveal new and conserved regulatory strategies.
Science, 349, 2015
|
|
5O8B
 
 | | Difference-refined excited-state structure of rsEGFP2 1ps following 400nm-laser irradiation of the off-state. | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
5O89
 
 | | Crystal Structure of rsEGFP2 in the fluorescent on-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
5O8C
 
 | | Composite structure of rsEGFP2 1ps following 400nm-laser irradiation of the off-state. | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
5NP1
 
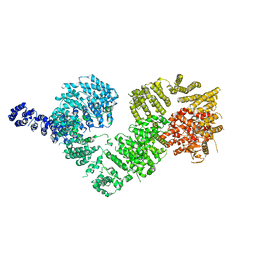 | | Open protomer of human ATM (Ataxia telangiectasia mutated) | | Descriptor: | Serine-protein kinase ATM | | Authors: | Baretic, D, Pollard, H.K, Fisher, D.I, Johnson, C.M, Santhanam, B, Truman, C.M, Kouba, T, Fersht, A.R, Phillips, C, Williams, R.L. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of closed and open conformations of dimeric human ATM.
Sci Adv, 3, 2017
|
|
8U57
 
 | | PPARg LBD in complex with perfluorooctanoic acid (PFOA) | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, Peroxisome proliferator-activated receptor gamma, pentadecafluorooctanoic acid | | Authors: | Pederick, J.L, Frkic, R.L, McDougal, D.P, Bruning, J.B. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural basis for the activation of peroxisome proliferator-activated receptor gamma (PPAR gamma ) by perfluorooctanoic acid (PFOA).
Chemosphere, 354, 2024
|
|
5KBK
 
 | | Candida Albicans Superoxide Dismutase 5 (SOD5), E110A Mutant | | Descriptor: | COPPER (I) ION, Cell surface Cu-only superoxide dismutase 5, SULFATE ION | | Authors: | Galaleldeen, A, Peterson, R.L, Villarreal, J, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-06-03 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.411 Å) | | Cite: | The Phylogeny and Active Site Design of Eukaryotic Copper-only Superoxide Dismutases.
J.Biol.Chem., 291, 2016
|
|
6D3E
 
 | | PPARg LBD in Complex with SR1988 | | Descriptor: | 1-[(2,4-difluorophenyl)methyl]-2,3-dimethyl-N-[(1R)-1-phenylpropyl]-1H-indole-5-carboxamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Frkic, R.L, Bruning, J.B. | | Deposit date: | 2018-04-15 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Structural and Dynamic Elucidation of a Non-acid PPARgammaPartial Agonist: SR1988.
Nucl Receptor Res, 5, 2018
|
|
4W8A
 
 | | Crystal structure of XEG5B, a GH5 xyloglucan-specific beta-1,4-glucanase from ruminal metagenomic library, in the native form | | Descriptor: | Exo-xyloglucanase, GLYCEROL, SULFATE ION | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
7ALX
 
 | | Sav-SOD: Chimeric Streptavidin-cSOD as Host for Artificial Metalloenzymes | | Descriptor: | ((4S)-1,3-dimesityl-4-((5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanamido)methyl)imidazolidin-2-yl)gold, Streptavidin,Superoxide dismutase [Cu-Zn],Streptavidin | | Authors: | Igareta, N.V, Christoffel, F, Peterson, R.L, Ward, T.R. | | Deposit date: | 2020-10-07 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and evolution of chimeric streptavidin for protein-enabled dual gold catalysis
Nat Catal, 4, 2021
|
|
3ZO1
 
 | | The Synthesis and Evaluation of Diazaspirocyclic Protein Kinase Inhibitors | | Descriptor: | 6-(1,9-DIAZASPIRO[5.5]UNDECAN-9-YL)-9H-PURINE, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, ... | | Authors: | Allen, C.E, Chow, C.L, Caldwell, J.J, Westwood, I.M, van Montfort, R.L, Collins, I. | | Deposit date: | 2013-02-20 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and evaluation of heteroaryl substituted diazaspirocycles as scaffolds to probe the ATP-binding site of protein kinases.
Bioorg. Med. Chem., 21, 2013
|
|
4IMM
 
 | |
4QX2
 
 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4W84
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in the native form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
6PEY
 
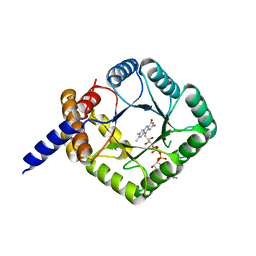 | | MTHFR with mutation Asp120Ala | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methylenetetrahydrofolate reductase | | Authors: | Gallagher, E.L, Gurney, L.A, Frasco, F.G, Trimmer, E, Bolen, R.L, Collins, K, Garland, E, Halloran, J, Handley-Pendelton, J, Hernandez, V, Leffler, S, Perez, A, Soares, A, Stojanoff, V, Williams, D. | | Deposit date: | 2019-06-21 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Examination of Asp120Ala a Chemically Important Novel Mutation in the Enzyme Mthylenetetrahydrofolate Reductase
To be published
|
|
6PKF
 
 | | Myocilin OLF mutant N428E/D478K | | Descriptor: | GLYCEROL, Myocilin | | Authors: | Lieberman, R.L, Hill, S.E. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Calcium-ligand variants of the myocilin olfactomedin propeller selected from invertebrate phyla reveal cross-talk with N-terminal blade and surface helices.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4W87
 
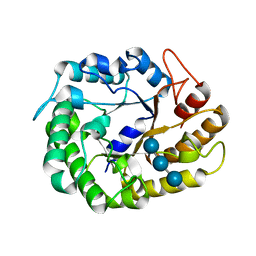 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from metagenomic library, in complex with a xyloglucan oligosaccharide | | Descriptor: | MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
4QX1
 
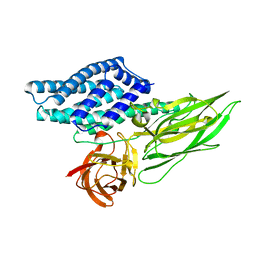 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the CrystFEL software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5D9Q
 
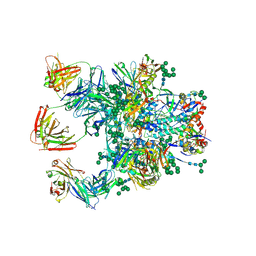 | | Crystal Structure of the BG505 SOSIP gp140 HIV-1 Env trimer in Complex with the Broadly Neutralizing Fab PGT122 and scFv NIH45-46 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Julien, J.-P, Stanfield, R.L, Ward, A.B, Wilson, I.A. | | Deposit date: | 2015-08-18 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Minimally Mutated HIV-1 Broadly Neutralizing Antibodies to Guide Reductionist Vaccine Design.
Plos Pathog., 12, 2016
|
|
5DHH
 
 | | The crystal structure of nociceptin/orphanin FQ peptide receptor (NOP) in complex with SB-612111 (PSI Community Target) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (5S,7S)-7-{[4-(2,6-dichlorophenyl)piperidin-1-yl]methyl}-1-methyl-6,7,8,9-tetrahydro-5H-benzo[7]annulen-5-ol, OLEIC ACID, ... | | Authors: | Miller, R.L, Thompson, A.A, Trapella, C, Guerrini, R, Malfacini, D, Patel, N, Han, G.W, Cherezov, V, Calo, G, Katritch, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2015-08-31 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | The Importance of Ligand-Receptor Conformational Pairs in Stabilization: Spotlight on the N/OFQ G Protein-Coupled Receptor.
Structure, 23, 2015
|
|
5DJ9
 
 | | Crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49 in a complex with gabaculine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-01 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49 in a complex with gabaculine
To Be Published
|
|
6PKD
 
 | | Myocilin OLF mutant N428D/D478H | | Descriptor: | Myocilin, SODIUM ION | | Authors: | Lieberman, R.L, Hill, S.E. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Calcium-ligand variants of the myocilin olfactomedin propeller selected from invertebrate phyla reveal cross-talk with N-terminal blade and surface helices.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
