4UEX
 
 | |
1YJD
 
 | | Crystal structure of human CD28 in complex with the Fab fragment of a mitogenic antibody (5.11A1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab fragment of 5.11A1 antibody heavy chain, Fab fragment of 5.11A1 antibody light chain, ... | | Authors: | Evans, E.J, Esnouf, R.M, Manso-Sancho, R, Gilbert, R.J.C, James, J.R, Sorensen, P, Stuart, D.I, Davis, S.J. | | Deposit date: | 2005-01-14 | | Release date: | 2005-02-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a soluble CD28-Fab complex
Nat.Immunol., 6, 2005
|
|
4UG3
 
 | | B. subtilis GpsB N-terminal Domain | | Descriptor: | CELL CYCLE PROTEIN GPSB | | Authors: | Rismondo, J, Cleverley, R.M, Lane, H.V, Grohennig, S, Steglich, A, Moller, L, Krishna Mannala, G, Hain, T, Lewis, R.J, Halbedel, S. | | Deposit date: | 2015-03-21 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Bacterial Cell Division Determinant Gpsb and its Interaction with Penicillin Binding Proteins.
Mol.Microbiol., 99, 2016
|
|
4UD4
 
 | | Structural Plasticity of Cid1 Provides a Basis for its RNA Terminal Uridylyl Transferase Activity | | Descriptor: | GLYCEROL, POLY(A) RNA POLYMERASE PROTEIN CID1 | | Authors: | Yates, L.A, Durrant, B.P, Fleurdepine, S, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | Deposit date: | 2014-12-07 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural plasticity of Cid1 provides a basis for its distributive RNA terminal uridylyl transferase activity.
Nucleic Acids Res., 43, 2015
|
|
4UD5
 
 | | Structural Plasticity of Cid1 Provides a Basis for its RNA Terminal Uridylyl Transferase Activity | | Descriptor: | POLY(A) RNA POLYMERASE PROTEIN CID1 | | Authors: | Yates, L.A, Durrant, B.P, Fleurdepine, S, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | Deposit date: | 2014-12-07 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural Plasticity of Cid1 Provides a Basis for its Distributive RNA Terminal Uridylyl Transferase Activity.
Nucleic Acids Res., 43, 2015
|
|
6QND
 
 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Xylose Isomerase 60 s timepoint | | Descriptor: | COBALT (II) ION, D-glucose, MAGNESIUM ION, ... | | Authors: | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | Deposit date: | 2019-02-11 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
6QNI
 
 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Xylose Isomerase 1.0 s timepoint | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, Xylose isomerase, ... | | Authors: | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | Deposit date: | 2019-02-11 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
6QN7
 
 | | Structure of bovine anti-RSV hybrid Fab B4HC-B13LC | | Descriptor: | Heavy chain of bovine anti-RSV B4, Light chain of bovine anti-RSV B13 | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
6QNH
 
 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Xylose Isomerase 0ms timepoint | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, Xylose isomerase | | Authors: | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | Deposit date: | 2019-02-11 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
6QNK
 
 | | Antibody FAB fragment targeting Gi protein heterotrimer | | Descriptor: | 1,2-ETHANEDIOL, D-MALATE, FAB heavy chain, ... | | Authors: | Tsai, C.-J, Muehle, J, Pamula, F, Dawson, R.J.P, Maeda, S, Deupi, X, Schertler, G.F.X. | | Deposit date: | 2019-02-11 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cryo-EM structure of the rhodopsin-G alpha i-beta gamma complex reveals binding of the rhodopsin C-terminal tail to the G beta subunit.
Elife, 8, 2019
|
|
4UFI
 
 | | Mouse Galactocerebrosidase complexed with aza-galacto-fagomine AGF | | Descriptor: | (3R,4S,5R)-3-(hydroxymethyl)-1,2-diazinane-4,5-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hill, C.H, Viuff, A.H, Spratley, S.J, Salamone, S, Christensen, S.H, Read, R.J, Moriarty, N.W, Jensen, H.H, Deane, J.E. | | Deposit date: | 2015-03-17 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Azasugar Inhibitors as Pharmacological Chaperones for Krabbe Disease.
Chem.Sci., 6, 2015
|
|
6RPZ
 
 | |
5S7N
 
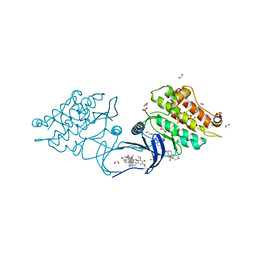 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010920a | | Descriptor: | 1,2-ETHANEDIOL, 1H-imidazol-2-amine, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S7W
 
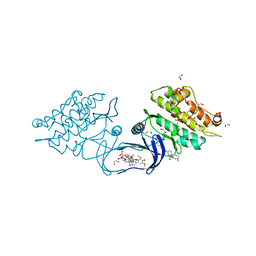 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with HM000007h | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type-1, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S7Y
 
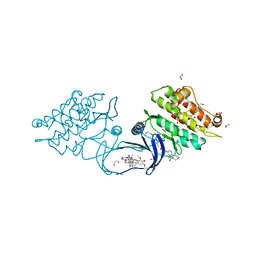 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010933a | | Descriptor: | (2S,4R)-1,3-thiazolidine-2,4-diamine, 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S84
 
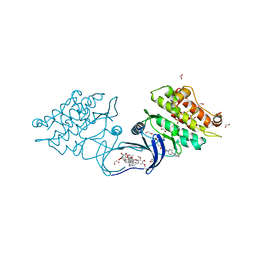 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010949a | | Descriptor: | 1,2-ETHANEDIOL, 1-[(4R)-1,3-oxazolidin-4-yl]methanamine, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S8A
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with NU074484b | | Descriptor: | (4S)-imidazolidine-4-carbonitrile, 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S78
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010934a | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type-1, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S7C
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM000274c | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type-1, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S7M
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM000275d | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type-1, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S7Q
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010944a | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, 5-methyl-1H-tetrazole, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S7P
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010937a | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type-1, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S85
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM000884c | | Descriptor: | (3R)-1,2-oxazolidin-3-amine, 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S7S
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010921a | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type-1, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S8B
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010960a | | Descriptor: | 1,2-ETHANEDIOL, 1lambda~6~-thietane-1,1-dione, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | XChem group deposition
To Be Published
|
|
