1HW1
 
 | | THE FADR-DNA COMPLEX: TRANSCRIPTIONAL CONTROL OF FATTY ACID METABOLISM IN ESCHERICHIA COLI | | Descriptor: | FATTY ACID METABOLISM REGULATOR PROTEIN, SULFATE ION, ZINC ION | | Authors: | Xu, Y, Heath, R.J, Li, Z, Rock, C.O, White, S.W. | | Deposit date: | 2001-01-09 | | Release date: | 2001-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The FadR.DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli.
J.Biol.Chem., 276, 2001
|
|
1HW2
 
 | | FADR-DNA COMPLEX: TRANSCRIPTIONAL CONTROL OF FATTY ACID METABOLISM IN ECHERICHIA COLI | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*TP*GP*GP*TP*CP*CP*GP*AP*CP*CP*AP*GP*AP*TP*GP*CP*T)-3', 5'-D(*G*CP*AP*TP*CP*TP*GP*GP*TP*CP*GP*GP*AP*CP*CP*AP*GP*AP*TP*CP*GP*A)-3', FATTY ACID METABOLISM REGULATOR PROTEIN, ... | | Authors: | Xu, Y, Heath, R.J, Li, Z, Rock, C.O, White, S.W. | | Deposit date: | 2001-01-09 | | Release date: | 2001-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The FadR.DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli.
J.Biol.Chem., 276, 2001
|
|
1HM3
 
 | | ACTIVE SITE OF CHONDROITINASE AC LYASE REVEALED BY THE STRUCTURE OF ENZYME-OLIGOSACCHARIDE COMPLEXES AND MUTAGENESIS | | Descriptor: | 2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, CALCIUM ION, CHONDROITINASE AC, ... | | Authors: | Huang, W, Boju, L, Tkalec, L, Su, H, Yang, H.O, Gunay, N.S, Linhardt, R.J, Kim, Y.S, Matte, A, Cygler, M. | | Deposit date: | 2000-12-04 | | Release date: | 2001-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis.
Biochemistry, 40, 2001
|
|
1HM2
 
 | | ACTIVE SITE OF CHONDROITINASE AC LYASE REVEALED BY THE STRUCTURE OF ENZYME-OLIGOSACCHARIDE COMPLEXES AND MUTAGENESIS | | Descriptor: | 2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, CALCIUM ION, CHONDROITINASE AC, ... | | Authors: | Huang, W, Boju, L, Tkalec, L, Su, H, Yang, H.O, Gunay, N.S, Linhardt, R.J, Kim, Y.S, Matte, A, Cygler, M. | | Deposit date: | 2000-12-04 | | Release date: | 2001-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis.
Biochemistry, 40, 2001
|
|
1IS9
 
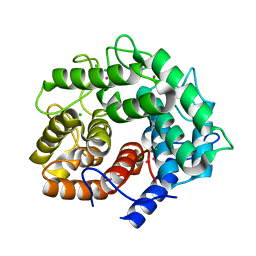 | | Endoglucanase A from Clostridium thermocellum at atomic resolution | | Descriptor: | CHLORIDE ION, MERCURY (II) ION, endoglucanase A | | Authors: | Schmidt, A, Gonzalez, A, Morris, R.J, Costabel, M, Alzari, P.M, Lamzin, V.S. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Advantages of high-resolution phasing: MAD to atomic resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4MIN
 
 | |
1FVT
 
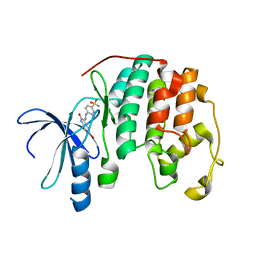 | | THE STRUCTURE OF CYCLIN-DEPENDENT KINASE 2 (CDK2) IN COMPLEX WITH AN OXINDOLE INHIBITOR | | Descriptor: | 4-[(2Z)-2-(5-bromo-2-oxo-1,2-dihydro-3H-indol-3-ylidene)hydrazinyl]benzene-1-sulfonamide, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Davis, S.T, Benson, B.G, Bramson, H.N, Chapman, D.E, Dickerson, S.H, Dold, K.M, Eberwein, D.J, Edelstein, M, Frye, S.V, Gampe Jr, R.T, Griffin, R.J, Harris, P.A, Hassell, A.M, Holmes, W.D, Hunter, R.N, Knick, V.B, Lackey, K, Lovejoy, B, Luzzio, M.J, Murray, D, Parker, P, Rocque, W.J, Shewchuk, L, Veal, J.M, Walker, D.H, Kuyper, L.K. | | Deposit date: | 2000-09-20 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prevention of chemotherapy-induced alopecia in rats by CDK inhibitors.
Science, 291, 2001
|
|
4MJL
 
 | | Crystal Structure of myo-inositol dehydrogenase from Lactobacillus casei in complex with NAD and D-chiro-inositol | | Descriptor: | (1R,2R,3S,4S,5S,6S)-CYCLOHEXANE-1,2,3,4,5,6-HEXOL, GLYCEROL, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, ... | | Authors: | Bertwistle, D, Sanders, D.A.R, Palmer, D.R.J. | | Deposit date: | 2013-09-03 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of myo-inositol dehydrogenase from Lactobacillus casei in complex with NAD and D-chiro-inositol
To be Published
|
|
1G5X
 
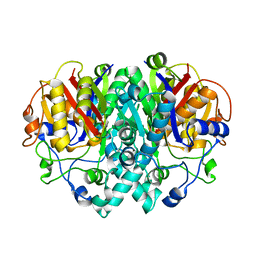 | | The Structure of Beta-Ketoacyl-[Acyl Carrier Protein] Synthase I | | Descriptor: | BETA-KETOACYL ACYL CARRIER PROTEIN SYNTHASE I | | Authors: | Zhang, Y.M, Rao, M.S, Heath, R.J, Price, A.C, Olson, A.J, Rock, C.O, White, S.W. | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification and analysis of the acyl carrier protein (ACP) docking site on beta-ketoacyl-ACP synthase III.
J.Biol.Chem., 276, 2001
|
|
7KZ6
 
 | | Crystal structure of KabA from Bacillus cereus UW85 with bound cofactor PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
7KZ3
 
 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the internal aldimine | | Descriptor: | 1,2-ETHANEDIOL, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, SODIUM ION | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-09 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
7KZ5
 
 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the plp external aldimine adduct with kanosamine-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 3-deoxy-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-O-phosphono-alpha-D-gluco pyranose, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, ... | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
7KZD
 
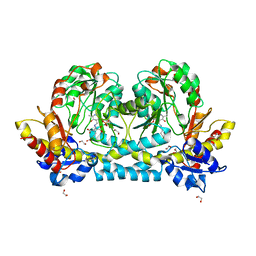 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the reduced internal aldimine and with bound Glutarate | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 1,2-ETHANEDIOL, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, ... | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
1FFH
 
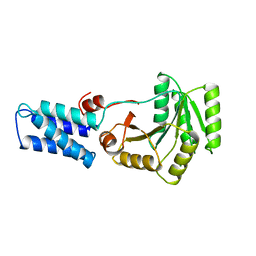 | | N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION PROTEIN FFH FROM THERMUS AQUATICUS | | Descriptor: | FFH, MAGNESIUM ION | | Authors: | Freymann, D.M, Keenan, R.J, Stroud, R.M, Walter, P. | | Deposit date: | 1996-12-30 | | Release date: | 1997-12-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the conserved GTPase domain of the signal recognition particle.
Nature, 385, 1997
|
|
1FDL
 
 | |
1G80
 
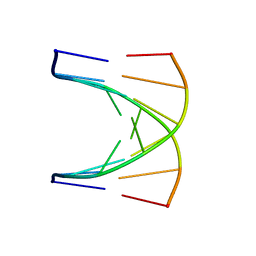 | | NMR SOLUTION STRUCTURE OF D(GCGTACGC)2 | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3' | | Authors: | Isaacs, R.J, Spielmann, H.P. | | Deposit date: | 2000-11-15 | | Release date: | 2001-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Relationship of DNA structure to internal dynamics: correlation of helical parameters from NOE-based NMR solution structures of d(GCGTACGC)(2) and d(CGCTAGCG)(2) with (13)C order parameters implies conformational coupling in dinucleotide units.
J.Mol.Biol., 307, 2001
|
|
1JKY
 
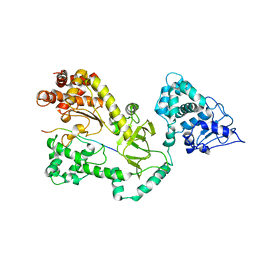 | | Crystal Structure of the Anthrax Lethal Factor (LF): Wild-type LF Complexed with the N-terminal Sequence of MAPKK2 | | Descriptor: | Lethal Factor, mitogen-activated protein kinase kinase 2 | | Authors: | Pannifer, A.D, Wong, T.Y, Schwarzenbacher, R, Renatus, M, Petosa, C, Collier, R.J, Bienkowska, J, Lacy, D.B, Park, S, Leppla, S.H, Hanna, P, Liddington, R.C. | | Deposit date: | 2001-07-13 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of the anthrax lethal factor.
Nature, 414, 2001
|
|
7LQS
 
 | | Structure of truncated conotoxin CIC | | Descriptor: | Alpha-conotoxin CIC | | Authors: | Evans, E.R.J, Daly, N.L. | | Deposit date: | 2021-02-15 | | Release date: | 2021-04-21 | | Method: | SOLUTION NMR | | Cite: | Synthesis, Structural and Pharmacological Characterizations of CIC, a Novel alpha-Conotoxin with an Extended N-Terminal Tail.
Mar Drugs, 19, 2021
|
|
4MIY
 
 | | Crystal Structure of myo-inositol dehydrogenase from Lactobacillus casei in complex with NAD and myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, GLYCEROL, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, ... | | Authors: | Bertwistle, D, Sanders, D.A.R, Palmer, D.R.J. | | Deposit date: | 2013-09-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of myo-inositol dehydrogenase from Lactobacillus casei in complex with NAD and myo-inositol
To be Published
|
|
4N54
 
 | | Crystal structure of scyllo-inositol dehydrogenase from Lactobacillus casei with bound cofactor NAD(H) and scyllo-inositol | | Descriptor: | (1r,2r,3r,4r,5r,6r)-cyclohexane-1,2,3,4,5,6-hexol, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Inositol dehydrogenase | | Authors: | Bertwistle, D, Sanders, D.A.R, Palmer, D.R.J. | | Deposit date: | 2013-10-09 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of scyllo-inositol dehydrogenase from Lactobacillus casei with bound cofactor NAD(H) and scyllo-inositol
To be Published
|
|
1HQL
 
 | | The xenograft antigen in complex with the B4 isolectin of Griffonia simplicifolia lectin-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LECTIN, ... | | Authors: | Tempel, W, Lipscomb, L.A, Rose, J.P, Woods, R.J. | | Deposit date: | 2000-12-18 | | Release date: | 2002-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The xenograft antigen bound to Griffonia simplicifolia lectin 1-B(4). X-ray crystal structure of the complex and molecular dynamics characterization of the binding site.
J.Biol.Chem., 277, 2002
|
|
2CME
 
 | | The crystal structure of SARS coronavirus ORF-9b protein | | Descriptor: | DECANE, HYPOTHETICAL PROTEIN 5 | | Authors: | Meier, C, Aricescu, A.R, Assenberg, R, Aplin, R.T, Gilbert, R.J.C, Grimes, J.M, Stuart, D.I. | | Deposit date: | 2006-05-06 | | Release date: | 2006-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of Orf-9B, a Lipid Binding Protein from the Sars Coronavirus.
Structure, 14, 2006
|
|
1FVV
 
 | | THE STRUCTURE OF CDK2/CYCLIN A IN COMPLEX WITH AN OXINDOLE INHIBITOR | | Descriptor: | 4-[(7-OXO-7H-THIAZOLO[5,4-E]INDOL-8-YLMETHYL)-AMINO]-N-PYRIDIN-2-YL-BENZENESULFONAMIDE, CYCLIN A, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Davis, S.T, Benson, B.G, Bramson, H.N, Chapman, D.E, Dickerson, S.H, Dold, K.M, Eberwein, D.J, Edelstein, M, Frye, S.V, Gampe Jr, R.T, Griffin, R.J, Harris, P.A, Hassell, A.M, Holmes, W.D, Hunter, R.N, Knick, V.B, Lackey, K, Lovejoy, B, Luzzio, M.J, Murray, D, Parker, P, Rocque, W.J, Shewchuk, L, Veal, J.M, Walker, D.H, Kuyper, L.K. | | Deposit date: | 2000-09-20 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Prevention of chemotherapy-induced alopecia in rats by CDK inhibitors.
Science, 291, 2001
|
|
1G7Z
 
 | | NMR SOLUTION STRUCTURE OF D(CGCTAGCG)2 | | Descriptor: | 5'-D(*CP*GP*CP*TP*AP*GP*CP*G)-3' | | Authors: | Isaacs, R.J, Spielmann, H.P. | | Deposit date: | 2000-11-15 | | Release date: | 2001-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Relationship of DNA structure to internal dynamics: correlation of helical parameters from NOE-based NMR solution structures of d(GCGTACGC)(2) and d(CGCTAGCG)(2) with (13)C order parameters implies conformational coupling in dinucleotide units.
J.Mol.Biol., 307, 2001
|
|
2CL8
 
 | | Dectin-1 in complex with beta-glucan | | Descriptor: | CALCIUM ION, CHLORIDE ION, DECTIN-1, ... | | Authors: | Brown, J, O'Callaghan, C.A, Marshall, A.S.J, Gilbert, R.J.C, Siebold, C, Gordon, S, Brown, G.D, Jones, E.Y. | | Deposit date: | 2006-04-26 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Fungal Beta-Glucan-Binding Immune Receptor Dectin-1: Implications for Function.
Protein Sci., 16, 2007
|
|
