3P9Z
 
 | | Crystal structure of uroporphyrinogen-III synthetase from Helicobacter pylori 26695 | | Descriptor: | MALONATE ION, Uroporphyrinogen III cosynthase (HemD) | | Authors: | Nocek, B, Stein, A, Chhor, G, Fenske, R.J, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-10-18 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of uroporphyrinogen-III synthetase from Helicobacter pylori 26695
To be Published
|
|
5JIF
 
 | | Crystal structure of mouse hepatitis virus strain DVIM Hemagglutinin-Esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zeng, Q.H, Bakkers, M.J.G, Feitsma, L.J, de Groot, R.J, Huizinga, E.G. | | Deposit date: | 2016-04-22 | | Release date: | 2016-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coronavirus receptor switch explained from the stereochemistry of protein-carbohydrate interactions and a single mutation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4L9X
 
 | | Triazine hydrolase from Arthobacter aurescens modified for maximum expression in E.coli | | Descriptor: | ACETATE ION, Triazine hydrolase | | Authors: | Jackson, C.J, Coppin, C.W, Alexandrov, A, Wilding, M, Liu, J.-W, Ubels, J, Paks, M, Carr, P.D, Newman, J, Russell, R.J, Field, M, Weik, M, Oakeshott, J.G, Scott, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 300-Fold increase in production of the Zn2+-dependent dechlorinase TrzN in soluble form via apoenzyme stabilization.
Appl.Environ.Microbiol., 80, 2014
|
|
3ZXL
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HIAXHD3 | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4A5M
 
 | | Redox regulator HypR in its oxidized form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UNCHARACTERIZED HTH-TYPE TRANSCRIPTIONAL REGULATOR YYBR | | Authors: | Palm, G.J, Waack, P, Read, R.J, Hinrichs, W. | | Deposit date: | 2011-10-26 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights Into the Redox-Switch Mechanism of the Marr/Duf24-Type Regulator Hypr
Nucleic Acids Res., 40, 2012
|
|
4AD1
 
 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens | | Descriptor: | GLYCOSYL HYDROLASE FAMILY 71 | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3OKL
 
 | | Crystal structure of S25-39 in complex with Kdo(2.8)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-25 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
4AD3
 
 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with Glucose-1,3-deoxymannojirimycin | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, GLYCOSYL HYDROLASE FAMILY 71, alpha-D-glucopyranose | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AKC
 
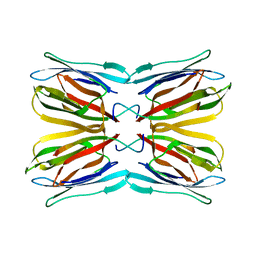 | | Structure of Galactose Binding lectin from Champedak (CGB) with Gal(beta)1,3-GalNac | | Descriptor: | AGGLUTININ ALPHA CHAIN, AGGLUTININ BETA-4 CHAIN, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Gabrielsen, M, Abdul-Rahman, P.S, Othman, S, Hashim, O.H, Cogdell, R.J. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures and Binding Specificity of Galactose- and Mannose-Binding Lectins from Champedak: Differences from Jackfruit Lectins
Acta Crystallogr.,Sect.F, 70, 2014
|
|
1Y8Y
 
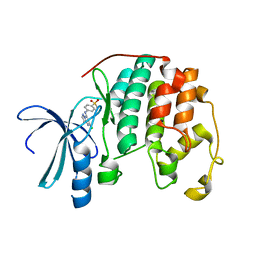 | | Crystal structure of human CDK2 complexed with a pyrazolo[1,5-a]pyrimidine inhibitor | | Descriptor: | (5-CHLOROPYRAZOLO[1,5-A]PYRIMIDIN-7-YL)-(4-METHANESULFONYLPHENYL)AMINE, Cell division protein kinase 2 | | Authors: | Williamson, D.S, Parratt, M.J, Torrance, C.J, Bower, J.F, Moore, J.D, Richardson, C.M, Dokurno, P, Cansfield, A.D, Francis, G.L, Hebdon, R.J, Howes, R, Jackson, P.S, Lockie, A.M, Murray, J.B, Nunns, C.L, Powles, J, Robertson, A, Surgenor, A.E. | | Deposit date: | 2004-12-14 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Structure-guided design of pyrazolo[1,5-a]pyrimidines as inhibitors of human cyclin-dependent kinase 2.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1Y91
 
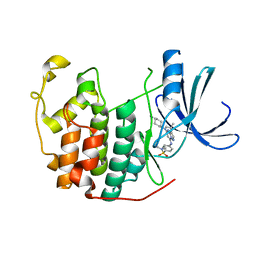 | | Crystal structure of human CDK2 complexed with a pyrazolo[1,5-a]pyrimidine inhibitor | | Descriptor: | 4-[5-(TRANS-4-AMINOCYCLOHEXYLAMINO)-3-ISOPROPYLPYRAZOLO[1,5-A]PYRIMIDIN-7-YLAMINO]-N,N-DIMETHYLBENZENESULFONAMIDE, Cell division protein kinase 2 | | Authors: | Williamson, D.S, Parratt, M.J, Torrance, C.J, Bower, J.F, Moore, J.D, Richardson, C.M, Dokurno, P, Cansfield, A.D, Francis, G.L, Hebdon, R.J, Howes, R, Jackson, P.S, Lockie, A.M, Murray, J.B, Nunns, C.L, Powles, J, Robertson, A, Surgenor, A.E. | | Deposit date: | 2004-12-14 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-guided design of pyrazolo[1,5-a]pyrimidines as inhibitors of human cyclin-dependent kinase 2.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
3ZFE
 
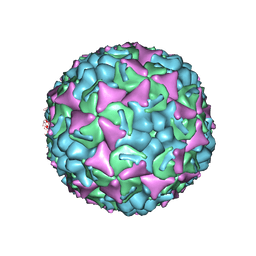 | | Human enterovirus 71 in complex with capsid binding inhibitor WIN51711 | | Descriptor: | CHLORIDE ION, SODIUM ION, SPHINGOSINE, ... | | Authors: | Plevka, P, Perera, R, Yap, M.L, Cardosa, J, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2012-12-11 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Human Enterovirus 71 in Complex with a Capsid-Binding Inhibitor.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5J69
 
 | |
4KQR
 
 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | Descriptor: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
3ZR6
 
 | | STRUCTURE OF GALACTOCEREBROSIDASE FROM MOUSE IN COMPLEX WITH GALACTOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Deane, J.E, Graham, S.C, Kim, N.N, Stein, P.E, Mcnair, R, Cachon-Gonzalez, M.B, Cox, T.M, Read, R.J. | | Deposit date: | 2011-06-14 | | Release date: | 2011-09-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Insights Into Krabbe Disease from Structures of Galactocerebrosidase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ZRZ
 
 | | Crystal structure of the second and third fibronectin F1 modules in complex with a fragment of Streptococcus pyogenes SfbI-5 | | Descriptor: | FIBRONECTIN, FIBRONECTIN-BINDING PROTEIN, GLYCEROL, ... | | Authors: | Norris, N.C, Bingham, R.J, Potts, J.R. | | Deposit date: | 2011-06-21 | | Release date: | 2011-09-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Analysis of the Tandem Beta-Zipper Interaction of a Streptococcal Protein with Human Fibronectin.
J.Biol.Chem., 286, 2011
|
|
1Y8M
 
 | |
1YFI
 
 | | Crystal Structure of restriction endonuclease MspI in complex with its cognate DNA in P212121 space group | | Descriptor: | 5'-D(*CP*CP*CP*CP*CP*GP*GP*GP*GP*G)-3', Type II restriction enzyme MspI | | Authors: | Xu, Q.S, Kucera, R.B, Roberts, R.J, Guo, H.-C. | | Deposit date: | 2005-01-02 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two crystal forms of the restriction enzyme MspI-DNA complex show the same novel structure.
Protein Sci., 14, 2005
|
|
3MMJ
 
 | | Structure of the PTP-like phytase from Selenomonas ruminantium in complex with myo-inositol hexakisphosphate | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2010-04-20 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate binding in protein-tyrosine phosphatase-like inositol polyphosphatases.
J.Biol.Chem., 287, 2012
|
|
3ZUD
 
 | | THERMOASCUS GH61 ISOZYME A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Otten, H, Quinlan, R.J, Sweeney, M.D, Poulsen, J.-C.N, Johansen, K.S, Krogh, K.B.R.M, Joergensen, C.I, Tovborg, M, Anthonsen, A, Tryfona, T, Walter, C.P, Dupree, P, Xu, F, Davies, G.J, Walton, P.H, Lo Leggio, L. | | Deposit date: | 2011-07-18 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Insights Into the Oxidative Degradation of Cellulose by a Copper Metalloenzyme that Exploits Biomass Components.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4MYF
 
 | | Crystal structure of Trypanosoma cruzi formiminoglutamase(oxidized) with Mn2+2 at pH 6.0 | | Descriptor: | Formiminoglutamase, MANGANESE (II) ION | | Authors: | Hai, Y, Dugery, R.J, Healy, D, Christianson, D.W. | | Deposit date: | 2013-09-27 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Formiminoglutamase from trypanosoma cruzi is an arginase-like manganese metalloenzyme.
Biochemistry, 52, 2013
|
|
4MYK
 
 | | Crystal structure of Trypanosoma cruzi formiminoglutamase (oxidized) with Mn2+2 at pH 8.5 | | Descriptor: | Formiminoglutamase, MANGANESE (II) ION | | Authors: | Hai, Y, Dugery, R.J, Healy, D, Christianson, D.W. | | Deposit date: | 2013-09-27 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.518 Å) | | Cite: | Formiminoglutamase from trypanosoma cruzi is an arginase-like manganese metalloenzyme.
Biochemistry, 52, 2013
|
|
3ZQ6
 
 | | ADP-ALF4 COMPLEX OF M. THERM. TRC40 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Sherrill, J, Mariappan, M, Dominik, P, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2011-06-08 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.107 Å) | | Cite: | A Conserved Archaeal Pathway for Tail-Anchored Membrane Protein Insertion.
Traffic, 12, 2011
|
|
4N5C
 
 | | Crystal structure of Ypp1 | | Descriptor: | Cargo-transport protein YPP1 | | Authors: | Wu, X, Chi, R.J, Baskin, J.M, Lucast, L, Burd, C.G, De Camilli, P, Reinisch, K.M. | | Deposit date: | 2013-10-09 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural insights into assembly and regulation of the plasma membrane phosphatidylinositol 4-kinase complex.
Dev.Cell, 28, 2014
|
|
3ME4
 
 | | Crystal structure of mouse RANK | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Walter, S.W, Liu, C, Zhu, X, Wu, Y, Owens, R.J, Stuart, D.I, Gao, B, Ren, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-06-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and Functional Insights of RANKL-RANK Interaction and Signaling.
J.Immunol., 2010
|
|
