6BAT
 
 | | Crystal Structure of Wild-Type GltPh in complex with L-aspartate | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Font, J, Scopelliti, A.J, Vandenberg, R.J, Boudker, O, Ryan, R.M. | | Deposit date: | 2017-10-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural characterisation reveals insights into substrate recognition by the glutamine transporter ASCT2/SLC1A5.
Nat Commun, 9, 2018
|
|
4HFH
 
 | | The GLIC pentameric Ligand-Gated Ion Channel (wild-type) complexed to bromoform | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Howard, R.J, Malherbe, L, Lee, U.S, Corringer, P.J, Harris, R.A, Delarue, M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for potentiation by alcohols and anaesthetics in a ligand-gated ion channel.
Nat Commun, 4, 2013
|
|
3CL0
 
 | | N1 Neuraminidase H274Y + oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, Neuraminidase | | Authors: | Collins, P, Haire, L.F, Lin, Y.P, Liu, J, Russell, R.J, Walker, P.A, Skehel, J.J, Martin, S.R, Hay, A.J, Gamblin, S.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants.
Nature, 453, 2008
|
|
6BDY
 
 | |
3CNZ
 
 | | Structural dynamics of the microtubule binding and regulatory elements in the kinesin-like calmodulin binding protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin heavy chain-like protein, MAGNESIUM ION | | Authors: | Vinogradova, M.V, Malanina, G.G, Reddy, V, Reddy, A.S.N, Fletterick, R.J. | | Deposit date: | 2008-03-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural dynamics of the microtubule binding and regulatory elements in the kinesin-like calmodulin binding protein.
J.Struct.Biol., 163, 2008
|
|
7R6R
 
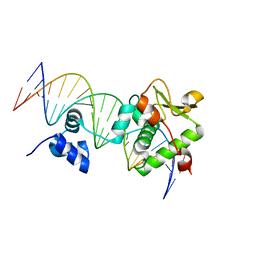 | | Crystal Structure of a Mycobacteriophage Cluster A2 Immunity Repressor:DNA Complex | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*TP*TP*GP*AP*CP*AP*GP*CP*CP*AP*CP*CP*GP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*CP*GP*GP*TP*GP*GP*CP*TP*GP*TP*CP*AP*AP*GP*CP*GP*GP*G)-3'), Immunity repressor | | Authors: | McGinnis, R.J, Brambley, C.A, Stamey, B, Green, W.C, Gragg, K.N, Cafferty, E.R, Terwilliger, T.C, Hammel, M, Hollis, T.J, Miller, J.M, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun, 13, 2022
|
|
3DGG
 
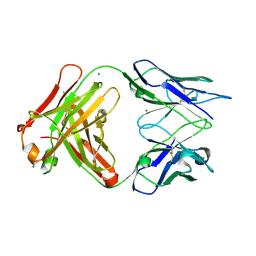 | | Crystal structure of FabOX108 | | Descriptor: | FabOX108 Heavy Chain Fragment, FabOX108 Light Chain Fragment, MAGNESIUM ION | | Authors: | Ren, J, Nettleship, J.E, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2008-06-13 | | Release date: | 2008-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A pipeline for the production of antibody fragments for structural studies using transient expression in HEK 293T cells.
Protein Expr.Purif., 62, 2008
|
|
3DV4
 
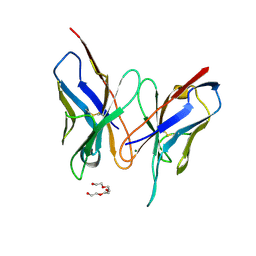 | | Crystal structure of SAG506-01, tetragonal, crystal 1 | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, Ig-like protein, MAGNESIUM ION, ... | | Authors: | Brooks, C.L, Blackler, R.J, Gerstenbruch, S, Kosma, P, Muller-Loennies, S, Brade, H, Evans, S.V. | | Deposit date: | 2008-07-18 | | Release date: | 2008-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pseudo-symmetry and twinning in crystals of homologous antibody Fv fragments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3DH9
 
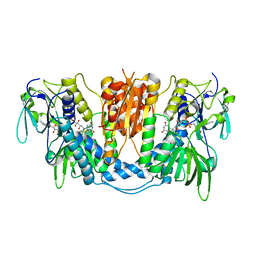 | |
8DSN
 
 | | Peptidylglycine alpha hydroxylating monoxygenase, Q272A | | Descriptor: | COPPER (II) ION, GLYCEROL, Peptidylglycine alpha-amidating monooxygenase | | Authors: | Arias, R.J, Blackburn, N.J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | New structures reveal flexible dynamics between the subdomains of peptidylglycine monooxygenase. Implications for an open to closed mechanism.
Protein Sci., 32, 2023
|
|
8DSJ
 
 | | Peptidylglycine alpha hydroxylating monooxygenase anaerobic | | Descriptor: | COPPER (II) ION, GLYCEROL, Peptidylglycine alpha-amidating monooxygenase | | Authors: | Arias, R.J, Blackburn, N.J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | New structures reveal flexible dynamics between the subdomains of peptidylglycine monooxygenase. Implications for an open to closed mechanism.
Protein Sci., 32, 2023
|
|
8DSL
 
 | | Peptidylglycine alpha hydroxylating monooxygenase, Q272E | | Descriptor: | COPPER (II) ION, GLYCEROL, Peptidylglycine alpha-amidating monooxygenase | | Authors: | Arias, R.J, Blackburn, N.J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | New structures reveal flexible dynamics between the subdomains of peptidylglycine monooxygenase. Implications for an open to closed mechanism.
Protein Sci., 32, 2023
|
|
3D1H
 
 | |
3D6R
 
 | |
6BAV
 
 | | Crystal Structure of GltPh R397C in complex with S-Benzyl-L-Cysteine | | Descriptor: | BENZYLCYSTEINE, Glutamate transporter homolog, SODIUM ION | | Authors: | Font, J, Scopelliti, A.J, Vandenberg, R.J, Boudker, O, Ryan, R.M. | | Deposit date: | 2017-10-16 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural characterisation reveals insights into substrate recognition by the glutamine transporter ASCT2/SLC1A5.
Nat Commun, 9, 2018
|
|
6BM5
 
 | |
3DIF
 
 | |
6BM6
 
 | |
3DUR
 
 | | Crystal structure of SAG173-04 | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, Ig-like protein, MAGNESIUM ION, ... | | Authors: | Brooks, C.L, Blackler, R.J, Gerstenbruch, S, Kosma, P, Muller-Loennies, S, Brade, H, Evans, S.V. | | Deposit date: | 2008-07-17 | | Release date: | 2008-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Pseudo-symmetry and twinning in crystals of homologous antibody Fv fragments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4J09
 
 | | Crystal Structure of LpxA bound to RJPXD33 | | Descriptor: | 1,2-ETHANEDIOL, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, ... | | Authors: | Jenkins, R.J, Meagher, J.L, Stuckey, J.A, Dotson, G.D. | | Deposit date: | 2013-01-30 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Recognition of Peptide RJPXD33 by Acyltransferases in Lipid A Biosynthesis.
J.Biol.Chem., 289, 2014
|
|
6BR6
 
 | | N2 neuraminidase in complex with a novel antiviral compound | | Descriptor: | (1R)-4-acetamido-3-amino-1,5-anhydro-2,3,4-trideoxy-1-sulfo-D-glycero-D-galacto-octitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, L.H, Ve, T, Pascolutti, M, Hadhazi, A, Bailly, B, Thomson, R.J, Gao, G.F, von Itzstein, M. | | Deposit date: | 2017-11-30 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.03983879 Å) | | Cite: | A Sulfonozanamivir Analogue Has Potent Anti-influenza Virus Activity.
ChemMedChem, 13, 2018
|
|
6BXA
 
 | | Crystal structure of N-terminal fragment of Zebrafish Toll-Like Receptor 5 (TLR5) with Lamprey Variable Lymphocyte Receptor 2 (VLR2) bound | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gunn, R.J, Wilson, I.A, Cooper, M.D, Herrin, B.R. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | VLR Recognition of TLR5 Expands the Molecular Characterization of Protein Antigen Binding by Non-Ig-based Antibodies.
J. Mol. Biol., 430, 2018
|
|
3G3Z
 
 | | The structure of NMB1585, a MarR family regulator from Neisseria meningitidis | | Descriptor: | Transcriptional regulator, MarR family | | Authors: | Nichols, C.E, Sainsbury, S, Ren, J, Walter, T.S, Verma, A, Stammers, D.K, Saunders, N.J, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-02-03 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of NMB1585, a MarR-family regulator from Neisseria meningitidis
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1VFB
 
 | |
7S6G
 
 | | Crystal structure of PhnD from Synechococcus MITS9220 in complex with phosphate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Shah, B.S, Mikolajek, H, Orr, C.M, Mykhaylyk, V, Owens, R.J, Paulsen, I.T. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Marine picocyanobacterial PhnD1 shows specificity for various phosphorus sources but likely represents a constitutive inorganic phosphate transporter.
Isme J, 17, 2023
|
|
