1RND
 
 | | NEWLY OBSERVED BINDING MODE IN PANCREATIC RIBONUCLEASE | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, RIBONUCLEASE A, SULFATE ION | | Authors: | Aguilar, C.F, Thomas, P.J, Mills, A, Moss, D.S, Palmer, R.A. | | Deposit date: | 1991-10-21 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Newly observed binding mode in pancreatic ribonuclease.
J.Mol.Biol., 224, 1992
|
|
6HQ6
 
 | | Bacterial beta-1,3-oligosaccharide phosphorylase from GH149 | | Descriptor: | 1,2-ETHANEDIOL, BICINE, Bacterial beta-1,3-oligosaccharide phosphorylase, ... | | Authors: | Kuhaudomlarp, S, Stevenson, C.E.M, Lawson, D.M, Field, R.A. | | Deposit date: | 2018-09-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of a GH149 beta-(1 → 3) glucan phosphorylase reveals a new surface oligosaccharide binding site and additional domains that are absent in the disaccharide-specific GH94 glucose-beta-(1 → 3)-glucose (laminaribiose) phosphorylase.
Proteins, 87, 2019
|
|
5FQR
 
 | | Selective estrogen receptor downregulator antagonists: Tetrahydroisoquinoline phenols 2. | | Descriptor: | (E)-3-[4-[(1R)-6-HYDROXY-2-ISOBUTYL-3,4-DIHYDRO-1H-ISOQUINOLIN-1-YL]PHENYL]PROP-2-ENOIC ACID, ESTROGEN RECEPTOR | | Authors: | Scott, J.S, Bailey, A, Davies, R.D.M, Degorce, S.L, MacFaul, P.A, Gingell, H, Moss, T, Norman, R.A, Pink, J.H, Rabow, A.A, Roberts, B, Smith, P.D. | | Deposit date: | 2015-12-14 | | Release date: | 2016-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Tetrahydroisoquinoline Phenols: Selective Estrogen Receptor Downregulator Antagonists with Oral Bioavailability in Rat.
Acs Med.Chem.Lett., 7, 2016
|
|
1XR6
 
 | | Crystal Structure of RNA-dependent RNA Polymerase 3D from human rhinovirus serotype 1B | | Descriptor: | Genome polyprotein, POTASSIUM ION | | Authors: | Love, R.A, Maegley, K.A, Yu, X, Ferre, R.A, Lingardo, L.K, Diehl, W, Parge, H.E, Dragovich, P.S, Fuhrman, S.A. | | Deposit date: | 2004-10-13 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of the RNA-Dependent RNA Polymerase from Human Rhinovirus: A Dual-Function Target for Common Cold Antiviral Therapy
Structure, 12, 2004
|
|
5FQV
 
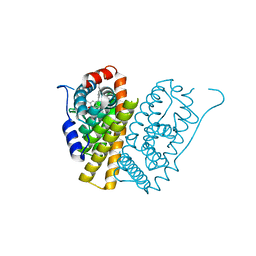 | | Selective estrogen receptor downregulator antagonists: Tetrahydroisoquinoline phenols 5. | | Descriptor: | (E)-3-[4-(6-hydroxy-2-isobutyl-7-methyl-3,4-dihydro-1H-isoquinolin-1-yl)phenyl]prop-2-enoic acid, ESTROGEN RECEPTOR ALPHA | | Authors: | Scott, J.S, bailey, A, Davies, R.D.M, Degorce, S.L, MacFaul, P.A, Gingell, H, Moss, T, Norman, R.A, Pink, J.H, Rabow, A.A, Roberts, B, Smith, P.D. | | Deposit date: | 2015-12-14 | | Release date: | 2016-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Tetrahydroisoquinoline Phenols: Selective Estrogen Receptor Downregulator Antagonists with Oral Bioavailability in Rat.
Acs Med.Chem.Lett., 7, 2016
|
|
7LHB
 
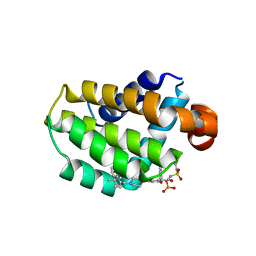 | | Crystal structure of Bcl-2 in complex with prodrug ABBV-167 | | Descriptor: | Apoptosis regulator Bcl-2, Phosphoric acid mono-[5-(5-{4-[2-(4-chloro-phenyl)-4,4-dimethyl-cyclohex-1-enylmethyl]-piperazin-1-yl}-2-{3-nitro-4-[(tetrahydro-pyran-4-ylmethyl)-amino]-benzenesulfonylaminocarbonyl}-phenoxy)-pyrrolo[2,3-b]pyridin-7-ylmethyl] ester | | Authors: | Judge, R.A, Salem, A.H. | | Deposit date: | 2021-01-21 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Expanding the Repertoire for "Large Small Molecules": Prodrug ABBV-167 Efficiently Converts to Venetoclax with Reduced Food Effect in Healthy Volunteers.
Mol.Cancer Ther., 20, 2021
|
|
2YIX
 
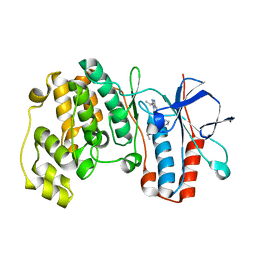 | | Triazolopyridine Inhibitors of p38 | | Descriptor: | 1-ethyl-3-(2-{[3-(1-methylethyl)[1,2,4]triazolo[4,3-a]pyridin-6-yl]sulfanyl}benzyl)urea, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Millan, D.S, Anderson, M, Bunnage, M.E, Burrows, J.L, Butcher, K.J, Dodd, P.G, Evans, T.J, Fairman, D.A, Han, s, Hughes, S.J, Irving, S.L, Kilty, I.C, Lemaitre, A, Lewthawaite, R.A, Mahke, A, Marr, E, Mathias, J.P, Philip, J, Phillips, C, Smith, R.T, Stefaniak, M.H, Yeadon, M. | | Deposit date: | 2011-05-17 | | Release date: | 2011-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Inhaled P38 Inhibitors for the Treatment of Chronic Obstructive Pulmonary Disease.
J.Med.Chem., 54, 2011
|
|
1XR7
 
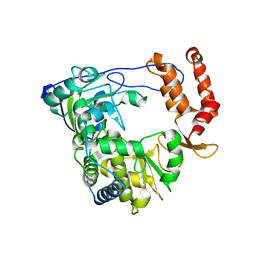 | | Crystal structure of RNA-dependent RNA Polymerase 3D from human rhinovirus serotype 16 | | Descriptor: | Genome polyprotein | | Authors: | Love, R.A, Maegley, K.A, Yu, X, Ferre, R.A, Lingardo, L.K, Diehl, W, Parge, H.E, Dragovich, P.S, Fuhrman, S.A. | | Deposit date: | 2004-10-13 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the RNA-Dependent RNA Polymerase from Human Rhinovirus: A Dual-Function Target for Common Cold Antiviral Therapy
Structure, 12, 2004
|
|
2WJ6
 
 | |
6HXF
 
 | | Human STK10 bound to a maleimide inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-methoxyphenyl)-4-[[4-(phenylcarbonyl)phenyl]amino]pyrrole-2,5-dione, CHLORIDE ION, ... | | Authors: | Sorrell, F.J, Salah, E, Serafim, R.A.M, Savitsky, P.A, Krojer, T, Bailey, H.J, Pinkas, D, Burgess-Brown, N.A, von Delft, F, Knapp, S, Arrowsmith, C, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2018-10-17 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Human STK10 bound to a maleimide inhibitor
To Be Published
|
|
1XR5
 
 | | Crystal Structure of the RNA-dependent RNA Polymerase 3D from human rhinovirus serotype 14 | | Descriptor: | Genome polyprotein, SAMARIUM (III) ION | | Authors: | Love, R.A, Maegley, K.A, Yu, X, Ferre, R.A, Lingardo, L.K, Diehl, W, Parge, H.E, Dragovich, P.S, Fuhrman, S.A. | | Deposit date: | 2004-10-13 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the RNA-Dependent RNA Polymerase from Human Rhinovirus: A Dual-Function Target for Common Cold Antiviral Therapy
Structure, 12, 2004
|
|
2WJ4
 
 | |
2WM2
 
 | |
7LZJ
 
 | |
3RN3
 
 | | SEGMENTED ANISOTROPIC REFINEMENT OF BOVINE RIBONUCLEASE A BY THE APPLICATION OF THE RIGID-BODY TLS MODEL | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Howlin, B, Moss, D.S, Harris, G.W, Palmer, R.A. | | Deposit date: | 1991-10-30 | | Release date: | 1991-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Segmented anisotropic refinement of bovine ribonuclease A by the application of the rigid-body TLS model.
Acta Crystallogr.,Sect.A, 45, 1989
|
|
1URW
 
 | | CDK2 IN COMPLEX WITH AN IMIDAZO[1,2-b]PYRIDAZINE | | Descriptor: | 2-[4-(N-(3-DIMETHYLAMINOPROPYL)SULPHAMOYL)ANILINO]-, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Byth, K.F, Cooper, N, Culshaw, J.D, Heaton, D.W, Oakes, S.E, Minshull, C.A, Norman, R.A, Pauptit, R.A, Tucker, J.A, Breed, J, Pannifer, A, Rowsell, S, Stanway, J.J, Valentine, A.L, Thomas, A.P. | | Deposit date: | 2003-11-11 | | Release date: | 2004-04-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Imidazo[1,2-B]Pyridazines: A Potent and Selective Class of Cyclin-Dependent Kinase Inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1UOU
 
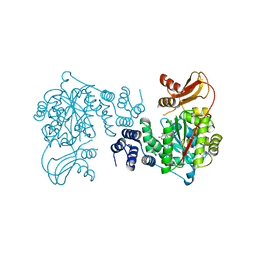 | | Crystal structure of human thymidine phosphorylase in complex with a small molecule inhibitor | | Descriptor: | 5-CHLORO-6-(1-(2-IMINOPYRROLIDINYL) METHYL) URACIL, THYMIDINE PHOSPHORYLASE | | Authors: | Norman, R.A, Barry, S.T, Bate, M, Breed, J, Colls, J.G, Ernill, R.J, Luke, R.W.A, Minshull, C.A, McAlister, M.S.B, McCall, E.J, McMiken, H.H.J, Paterson, D.S, Timms, D, Tucker, J.A, Pauptit, R.A. | | Deposit date: | 2003-09-23 | | Release date: | 2004-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal Structure of Human Thymidine Phosphorylase in Complex with a Small Molecule Inhibitor
Structure, 12, 2004
|
|
4I5P
 
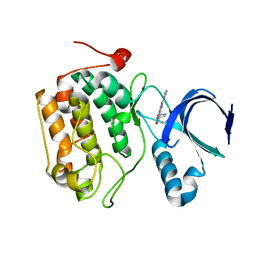 | | Selective & Brain-Permeable Polo-like Kinase-2 (Plk-2) Inhibitors that Reduce -Synuclein Phosphorylation in Rat Brain | | Descriptor: | (7R)-8-cyclopentyl-7-ethyl-5-methyl-2-(1H-pyrrol-2-yl)-7,8-dihydropteridin-6(5H)-one, Serine/threonine-protein kinase PLK2 | | Authors: | Aubele, D.L, Hom, R.K, Adler, M, Galemmo Jr, R.A, Bowers, S, Truong, A.P, Pan, H, Beroza, P. | | Deposit date: | 2012-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Selective and brain-permeable polo-like kinase-2 (Plk-2) inhibitors that reduce alpha-synuclein phosphorylation in rat brain.
Chemmedchem, 8, 2013
|
|
4I5M
 
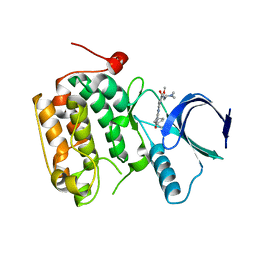 | | Selective & Brain-Permeable Polo-like Kinase-2 (Plk-2) Inhibitors that Reduce -Synuclein Phosphorylation in Rat Brain | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Serine/threonine-protein kinase PLK2 | | Authors: | Aubele, D.L, Hom, R.K, Adler, M, Galemmo Jr, R.A, Bowers, S, Truong, A.P, Pan, H, Beroza, P, Neitz, R.J, Yao, N, Lin, M, Tonn, G, Zhang, H, Bova, M.P, Ren, Z, Tam, D, Ruslim, L, Baker, J, Diep, L, Fitzgerald, K, Hoffman, J, Motter, R, Fauss, D, Tanaka, P, Dappen, M, Jagodzinski, J, Chan, W, Konradi, A.W, Latimer, L, Zhu, Y.L, Artis, D.R, Sham, H.L, Anderson, J.P, Bergeron, M. | | Deposit date: | 2012-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Selective and brain-permeable polo-like kinase-2 (Plk-2) inhibitors that reduce alpha-synuclein phosphorylation in rat brain.
Chemmedchem, 8, 2013
|
|
8G6L
 
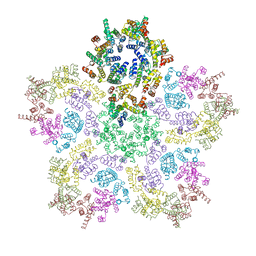 | | HIV-1 capsid lattice bound to IP6, pH 6.2 | | Descriptor: | Capsid protein | | Authors: | Highland, C.M, Dick, R.A. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2ACV
 
 | | Crystal Structure of Medicago truncatula UGT71G1 | | Descriptor: | URIDINE-5'-DIPHOSPHATE, triterpene UDP-glucosyl transferase UGT71G1 | | Authors: | Shao, H, He, X, Achnine, L, Blount, J.W, Dixon, R.A, Wang, X. | | Deposit date: | 2005-07-19 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of a Multifunctional Triterpene/Flavonoid Glycosyltransferase from Medicago truncatula.
Plant Cell, 17, 2005
|
|
8G6O
 
 | | HIV-1 capsid lattice bound to IP6 and Lenacapavir | | Descriptor: | Capsid protein | | Authors: | Highland, C.M, Dick, R.A. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G6K
 
 | |
8G6M
 
 | | HIV-1 CA lattice bound to IP6, pH 7.4 | | Descriptor: | Capsid protein | | Authors: | Highland, C.M, Dick, R.A. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G6N
 
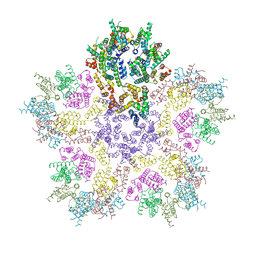 | | HIV-1 capsid lattice bound to dNTPs | | Descriptor: | Capsid protein | | Authors: | Highland, C.M, Dick, R.A. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
