5GGG
 
 | |
5GGN
 
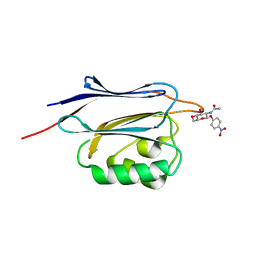 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GlcNAc-beta-pNP | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7XNX
 
 | |
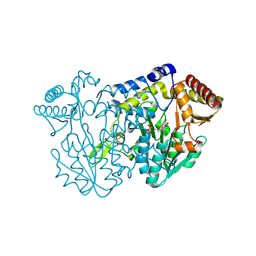
7XEK
 
 | | SufS with D-cysteine for 6 h | | Descriptor: | (2~{S},4~{S})-2-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Fujishiro, T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | X-ray crystallographic snapshots of the thioazolidine formation upon the PLP during inhibition of SufS by D-cysteine
to be published
|
|
7XEN
 
 | | SufS with L-penicillamine | | Descriptor: | (2~{R})-3-methyl-2-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-sulfanyl-butanoic acid, Cysteine desulfurase SufS | | Authors: | Nakamura, R, Fujishiro, T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | X-ray crystallographic snapshots of the thioazolidine formation upon the PLP during inhibition of SufS by D-cysteine
to be published
|
|
7XET
 
 | | SufS with beta-cyano-L-alanine | | Descriptor: | (2~{E})-3-cyano-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylimino]propanoic acid, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Fujishiro, T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibition mechanism of SufS by L-propargylglycine
to be published
|
|
7XEP
 
 | | SufS with L-propargylglycine | | Descriptor: | 1,2-ETHANEDIOL, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Fujishiro, T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Inhibition mechanism of SufS by L-propargylglycine
to be published
|
|
5GIT
 
 | | BTB domain of KEAP1 in complex with XX3 | | Descriptor: | Kelch-like ECH-associated protein 1, [(3aS,5R,5aS,6S,8S,8aS,9S,9aR)-9-acetyloxy-8-hydroxy-5,8a-dimethyl-1-methylidene-2-oxo-4,5,5a,6,7,8,9,9a-octahydro-3aH-azuleno[6,5-b]furan-6-yl] acetate | | Authors: | Zhu, L.L, Li, H.L, Wu, F.S, Xiong, R. | | Deposit date: | 2016-06-25 | | Release date: | 2017-06-28 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Britanin Ameliorates Cerebral Ischemia-Reperfusion Injury by Inducing the Nrf2 Protective Pathway.
Antioxid. Redox Signal., 27, 2017
|
|
7XEQ
 
 | | NifS with D-cysteine | | Descriptor: | (2~{S},4~{S})-2-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, Cysteine desulfurase IscS, ... | | Authors: | Nakamura, R, Fujishiro, T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray crystallographic snapshots of the thioazolidine formation upon the PLP during inhibition of SufS by D-cysteine
to be published
|
|
7XEL
 
 | | SufS soaked with D-penicillamine | | Descriptor: | 1,2-ETHANEDIOL, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Fujishiro, T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic snapshots of the thioazolidine formation upon the PLP during inhibition of SufS by D-cysteine
to be published
|
|
7MIZ
 
 | | Atomic structure of cortical microtubule from Toxoplasma gondii | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, X, Brown, A, Sibley, L.D, Zhang, R. | | Deposit date: | 2021-04-18 | | Release date: | 2021-06-02 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of cortical microtubules from human parasite Toxoplasma gondii identifies their microtubule inner proteins.
Nat Commun, 12, 2021
|
|
7VWS
 
 | | Carbazole Prenyl Transferase LvqB4 | | Descriptor: | 2-methyl-1-[(2R)-2-oxidanylpropyl]-9H-carbazole-3,4-dione, LvqB4, MAGNESIUM ION, ... | | Authors: | Suemune, H, Nagata, R, Kuzuyama, T, Nagano, S. | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural Basis for the Prenylation Reaction of Carbazole-Containing Natural Products Catalyzed by Squalene Synthase-Like Enzymes.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5E9Z
 
 | | Cytochrome P450 BM3 mutant M11 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Bifunctional cytochrome P450/NADPH--P450 reductase, FE (II) ION, ... | | Authors: | Capoferri, L, Leth, R, ter Haar, E, Mohanty, A.K, Grootenhuis, D.J, Vottero, E, Commandeur, J.N.M, Vermeulen, N.P.E, Jorgensen, F.S, Olsen, L, Geerke, D.P. | | Deposit date: | 2015-10-15 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Insights into regioselective metabolism of mefenamic acid by cytochrome P450 BM3 mutants through crystallography, docking, molecular dynamics, and free energy calculations.
Proteins, 84, 2016
|
|
7VWT
 
 | | Carbazole Prenyl Transferase CqsB4 | | Descriptor: | 2-methyl-1-[(2R)-2-oxidanylpropyl]-9H-carbazole-3,4-dione, CqsB4, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Suemune, H, Nagata, R, Kuzuyama, T, Nagano, S. | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis for the Prenylation Reaction of Carbazole-Containing Natural Products Catalyzed by Squalene Synthase-Like Enzymes.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5DK9
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a phenylamino-substituted ethyl triaryl-ethylene derivative 4,4'-{2-[3-(phenylamino)phenyl]but-1-ene-1,1-diyl}diphenol | | Descriptor: | 4,4'-{2-[3-(phenylamino)phenyl]but-1-ene-1,1-diyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-03 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
7MS7
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (5-((4-(4-chlorophenyl)piperidin-1-yl)sulfonyl)picolinoyl)glycine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, N-{5-[4-(4-chlorophenyl)piperidine-1-sulfonyl]pyridine-2-carbonyl}glycine, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7MS5
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 4-(4-(4-(3,4-difluoro-phenyl)-piperidin-1-ylsulfonyl)-phenyl)-4-oxo-butanoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[4-(3,4-difluorophenyl)piperidine-1-sulfonyl]phenyl}-4-oxobutanoic acid, CALCIUM ION, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
5E3E
 
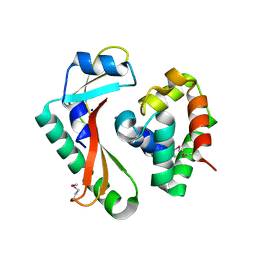 | | Crystal structure of CdiA-CT/CdiI complex from Y. kristensenii 33638 | | Descriptor: | CdiI immunity protein, Large exoprotein involved in heme utilization or adhesion, SODIUM ION | | Authors: | Michalska, K, Joachimiak, G, Jedrzejczak, R, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-10-02 | | Release date: | 2015-11-25 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The CDI toxin of Yersinia kristensenii is a novel bacterial member of the RNase A superfamily.
Nucleic Acids Res., 45, 2017
|
|
7MS6
 
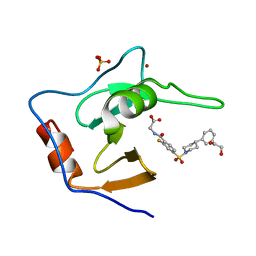 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (2-fluoro-4-((4-phenylpiperidin-1-yl)sulfonyl)benzoyl)glycine | | Descriptor: | 1,2-ETHANEDIOL, N-[2-fluoro-4-(4-phenylpiperidine-1-sulfonyl)benzoyl]glycine, SULFATE ION, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7VK9
 
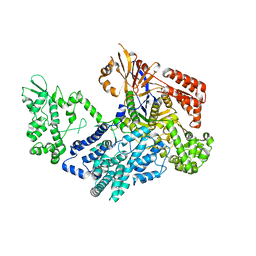 | | Crystal structure of xCas9 P411T | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1 | | Authors: | Bao, R, Liu, H.Y, Luo, Y.Z, Song, Y.J. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Dynamics Studies of the Spcas9 Variant Provide Insights into the Regulatory Role of the REC1 Domain
Acs Catalysis, 12, 2022
|
|
5DUN
 
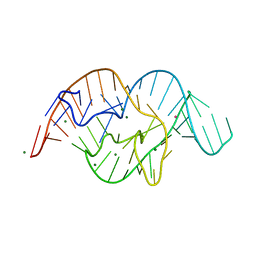 | | The crystal structure of OMe substituted twister ribozyme | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (54-MER) | | Authors: | Ren, A, Patel, D.J, Micura, R, Rajashankar, K.R. | | Deposit date: | 2015-09-19 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | A Mini-Twister Variant and Impact of Residues/Cations on the Phosphodiester Cleavage of this Ribozyme Class.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5DVM
 
 | | Fc Design 20.8.37 B chain homodimer E357D/S364R/Y407A | | Descriptor: | Fc-III peptide, Ig gamma-1 chain C region | | Authors: | Atwell, S, Leaver-Fay, A, Froning, K.J, Aldaz, H, Pustilnik, A, Lu, F, Huang, F, Yuan, R, Dhanani, S.H, Chamberlain, A.K, Fitchett, J.R, Gutierrez, B, Hendle, J, Demarest, S.J, Kuhlman, B. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-30 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Computationally Designed Bispecific Antibodies using Negative State Repertoires.
Structure, 24, 2016
|
|
5E5V
 
 | | Structure of amyloid forming peptide NFGAILS (residues 22-28) from Islet Amyloid Polypeptide | | Descriptor: | NFGAILS (22-28) from islet amyloid polypeptide, synthesized | | Authors: | Soriaga, A.B, Macdonald, R, Sawaya, M.R, Sangwan, S, Eisenberg, D. | | Deposit date: | 2015-10-09 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal Structures of IAPP Amyloidogenic Segments Reveal a Novel Packing Motif of Out-of-Register Beta Sheets.
J.Phys.Chem.B, 120, 2016
|
|
7V9P
 
 | |
7V9Q
 
 | |
