5XXM
 
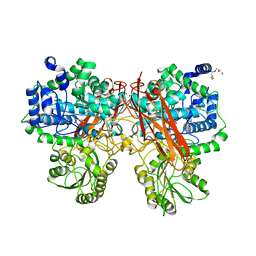 | | Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahash, Y, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
7WKP
 
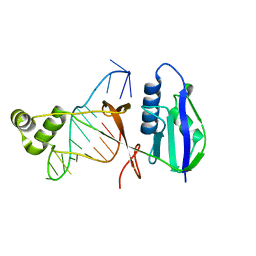 | | ICP1 Csy4 | | Descriptor: | 3' stem loop crRNA, Csy4 | | Authors: | Zhang, M, Peng, R. | | Deposit date: | 2022-01-10 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5XXW
 
 | | GDP-microtubule complexed with KIF5C in ATP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Morikawa, M, Shigematsu, H, Nitta, R, Hirokawa, N. | | Deposit date: | 2017-07-05 | | Release date: | 2018-10-10 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Kinesin-binding-triggered conformation switching of microtubules contributes to polarized transport
J. Cell Biol., 217, 2018
|
|
7TOG
 
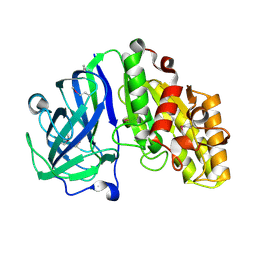 | | Crystal structure of carbohydrate esterase PbeAcXE, apoenzyme | | Descriptor: | SGNH hydrolase | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
6Y95
 
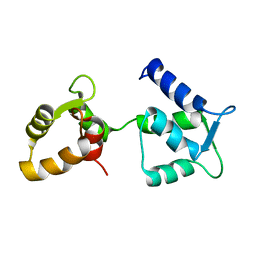 | | Ca2+-free Calmodulin mutant N53I | | Descriptor: | Calmodulin | | Authors: | Holt, C, Hamborg, L.N, Lau, K, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Petegem, F.V, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-03-06 | | Release date: | 2020-04-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
5XZE
 
 | | Mouse cGAS bound to the inhibitor RU332 | | Descriptor: | (3R)-3-[1-(3H-1lambda~4~,3-benzothiazol-2-yl)-5-hydroxy-3-methyl-1H-pyrazol-4-yl]-2-benzofuran-1(3H)-one, Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*G)-3'), ... | | Authors: | Vincent, J, Adura, C, Gao, P, Luz, A, Lama, L, Asano, Y, Okamoto, R, Imaeda, T, Aida, J, Rothamel, K, Gogakos, T, Steinberg, J, Reasoner, S, Aso, K, Tuschl, T, Patel, D.J, Glickman, J.F, Ascano, M. | | Deposit date: | 2017-07-12 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice.
Nat Commun, 8, 2017
|
|
7WKO
 
 | | ICP1 Csy1-2 complex | | Descriptor: | Csy1, Csy2 | | Authors: | Zhang, M, Peng, R. | | Deposit date: | 2022-01-10 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7TOI
 
 | | Crystal structure of carbohydrate esterase PbeAcXE, in complex with acetate | | Descriptor: | ACETATE ION, SGNH hydrolase | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
6Y3K
 
 | | NMR solution structure of the hazelnut allergen Cor a 1.0403 | | Descriptor: | Major allergen variant Cor a 1.0403 | | Authors: | Fuehrer, S, Kamenik, A.S, Zeindl, R, Nothegger, B, Hofer, F, Reider, N, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Sci Rep, 11, 2021
|
|
7TOJ
 
 | | Crystal structure of carbohydrate esterase CspAcXE, apoenzyme | | Descriptor: | CHLORIDE ION, SGNH/GDSL hydrolase family protein | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
5XXR
 
 | |
7TOH
 
 | | Crystal structure of carbohydrate esterase PbeAcXE, in complex with MeGlcpA-Xylp | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose, SGNH hydrolase | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
7P36
 
 | | X-ray structure of Lactobacillus kefir alcohol dehydrogenase (wild type) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bischoff, D, Walla, B, Janowski, R, Niessing, D, Weuster-Botz, D. | | Deposit date: | 2021-07-07 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Transfer of a Rational Crystal Contact Engineering Strategy between Diverse Alcohol Dehydrogenases
Crystals, 11, 2021
|
|
5AH4
 
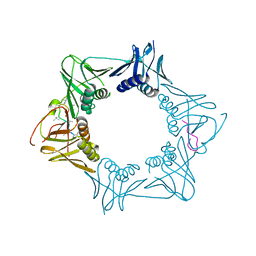 | | The sliding clamp of Mycobacterium smegmatis in complex with a natural product. | | Descriptor: | CYCLOHEXYL GRISELIMYCIN, DNA POLYMERASE III SUBUNIT BETA, SODIUM ION | | Authors: | Lukat, P, Kling, A, Heinz, D.W, Mueller, R. | | Deposit date: | 2015-02-04 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Antibiotics. Targeting Dnan for Tuberculosis Therapy Using Novel Griselimycins.
Science, 348, 2015
|
|
5XYX
 
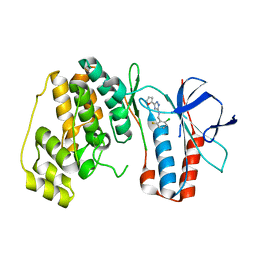 | | The structure of p38 alpha in complex with a triazol inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N-(2-chloro-6-fluorobenzyl)-5-(furan-2-yl)-2H-1,2,4-triazol-3-amine | | Authors: | Wang, Y.L, Sun, Y.Z, Cao, R, Liu, D, Li, L, Qi, X.B, Huang, N. | | Deposit date: | 2017-07-11 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | In Silico Identification of a Novel Hinge-Binding Scaffold for Kinase Inhibitor Discovery.
J. Med. Chem., 60, 2017
|
|
5A3U
 
 | | HIF prolyl hydroxylase 2 (PHD2/EGLN1) in complex with 6-(5-oxo-4-(1H- 1,2,3-triazol-1-yl)-2,5-dihydro-1H-pyrazol-1-yl)nicotinic acid | | Descriptor: | 6-(5-oxo-4-(1H-1,2,3-triazol-1-yl)-2,5-dihydro-1H-pyrazol-1-yl)nicotinic acid, EGL NINE HOMOLOG 1, MANGANESE (II) ION | | Authors: | Chowdhury, R, Gomez-Perez, V, Schofield, C.J. | | Deposit date: | 2015-06-03 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Potent and Selective Triazole-Based Inhibitors of the Hypoxia-Inducible Factor Prolyl-Hydroxylases with Activity in the Murine Brain.
Plos One, 10, 2015
|
|
6YTO
 
 | |
5A3P
 
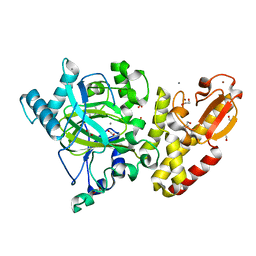 | | Crystal structure of the catalytic domain of human PLU1 (JARID1B). | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Nowak, R, Srikannathasan, V, Johansson, C, Gileadi, C, Tallant, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2015-06-02 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
6YU6
 
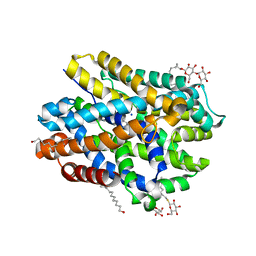 | | Crystal structure of MhsT in complex with L-leucine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, LEUCINE, SODIUM ION, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
7OF3
 
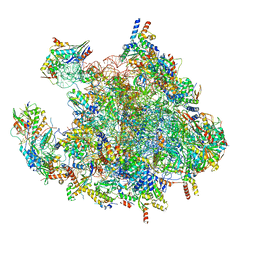 | | Structure of a human mitochondrial ribosome large subunit assembly intermediate in complex with MTERF4-NSUN4 (dataset2). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-05-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
5Y00
 
 | | Acid-tolerant monomeric GFP, Gamillus, fluorescence (ON) state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Sakurai, K, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2017-07-14 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Acid-Tolerant Monomeric GFP from Olindias formosa.
Cell Chem Biol, 25, 2018
|
|
4N63
 
 | |
7O25
 
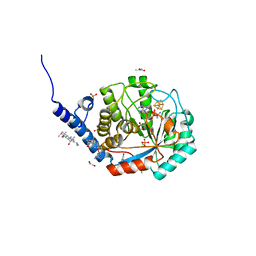 | | Complex-B bound [FeFe]-hydrogenase maturase HydE from T. maritima (reaction triggered in the crystal) | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBON MONOXIDE, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O1P
 
 | | [FeFe]-hydrogenase maturase HydE from T. Maritima (C-ter stretch absent) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O26
 
 | | Complex-B bound [FeFe]-hydrogenase maturase HydE fromT. Maritima (5'dA + Methionine) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 5'-DEOXYADENOSINE, CARBON MONOXIDE, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
