7AO5
 
 | | Crystal structure of CotB2 variant W288F | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclooctat-9-en-7-ol synthase, MAGNESIUM ION | | Authors: | Dimos, N, Driller, R, Loll, B. | | Deposit date: | 2020-10-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The Impression of a Nonexisting Catalytic Effect: The Role of CotB2 in Guiding the Complex Biosynthesis of Cyclooctat-9-en-7-ol.
J.Am.Chem.Soc., 142, 2020
|
|
7AO3
 
 | | Crystal structure of CotB2 variant F149L in complex with alendronate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, CHLORIDE ION, ... | | Authors: | Dimos, N, Driller, R, Loll, B. | | Deposit date: | 2020-10-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Impression of a Nonexisting Catalytic Effect: The Role of CotB2 in Guiding the Complex Biosynthesis of Cyclooctat-9-en-7-ol.
J.Am.Chem.Soc., 142, 2020
|
|
1FB5
 
 | | LOW RESOLUTION STRUCTURE OF OVINE ORNITHINE TRANSCARBMOYLASE IN THE UNLIGANDED STATE | | Descriptor: | NORVALINE, ORNITHINE TRANSCARBAMOYLASE | | Authors: | Zanotti, G, Battistutta, R, Panzalorto, M, Francescato, P, Bruno, G, De Gregorio, A. | | Deposit date: | 2000-07-14 | | Release date: | 2003-08-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Functional and structural characterization of ovine ornithine transcarbamoylase.
Org.Biomol.Chem., 1, 2003
|
|
5IY8
 
 | | Human holo-PIC in the initial transcribing state | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
7CX2
 
 | | Cryo-EM structure of the PGE2-bound EP2-Gs complex | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7AS4
 
 | | Recombinant human gTuRC | | Descriptor: | Actin, cytoplasmic 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Serna, M, Fernandez-Leiro, R, Llorca, O. | | Deposit date: | 2020-10-26 | | Release date: | 2021-01-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Assembly of the asymmetric human gamma-tubulin ring complex by RUVBL1-RUVBL2 AAA ATPase.
Sci Adv, 6, 2020
|
|
3F2I
 
 | | Crystal structure of the alr0221 protein from Nostoc, Northeast Structural Genomics Consortium Target NsR422. | | Descriptor: | Alr0221 protein, CHLORIDE ION, PHOSPHATE ION | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Foote, E.L, Ciccosanti, C, Belote, R.L, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-29 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the alr0221 protein from Nostoc, Northeast Structural Genomics Consortium Target NsR422.
To be Published
|
|
3F08
 
 | | Crystal structure of the putative uncharacterized protein Q6HG14 from Bacilllus thuringiensis. Northeast Structural Genomics Consortium target BuR153. | | Descriptor: | uncharacterized protein Q6HG14 | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Wang, H, Mao, L, Ciccosanti, C, Xiao, R, Nair, R, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-24 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the putative uncharacterized protein Q6HG14 from Bacilllus thuringiensis. Northeast Structural Genomics Consortium target BuR153.
To be Published
|
|
7B35
 
 | | MST3 in complex with compound MRIA13 | | Descriptor: | 8-[(5-azanyl-1,3-dioxan-2-yl)methyl]-6-[2-chloranyl-4-(3-methoxy-6-methyl-pyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.40005136 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
4XMV
 
 | |
7B6G
 
 | | Crystal structure of MurE from E.coli in complex with Z1675346324 | | Descriptor: | DIMETHYL SULFOXIDE, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase, trans-3-[(2,6-dimethylpyrimidin-4-yl)(methyl)amino]cyclobutan-1-ol | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7B6Q
 
 | | Crystal structure of MurE from E.coli in complex with Z57299526 | | Descriptor: | ISOPROPYL ALCOHOL, N-[(furan-2-yl)methyl]-1H-benzimidazol-2-amine, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
4XN1
 
 | |
5IXD
 
 | |
4XNB
 
 | |
5J7N
 
 | | Crystal structure of a small heat-shock protein from Xylella fastidiosa reveals a distinct high order structure | | Descriptor: | Low molecular weight heat shock protein | | Authors: | Fonseca, E.M.B, Scorsato, V, dos Santos, C.A, Tomazini Jr, A, Aparicio, R, Polikarpov, I. | | Deposit date: | 2016-04-06 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a small heat-shock protein from Xylella fastidiosa reveals a distinct high-order structure.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
4XO5
 
 | |
7B30
 
 | | MST3 in complex with compound G-5555 | | Descriptor: | 8-[(trans-5-amino-1,3-dioxan-2-yl)methyl]-6-[2-chloro-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, Serine/threonine-protein kinase 24 | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AJK
 
 | | Crystal structure of CRYI-B Rac1 complex | | Descriptor: | CYFIP-related Rac1 interactor B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Yelland, T, Anh, L, Insall, R, Machesky, L, Ismail, S. | | Deposit date: | 2020-09-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of CYRI-B Direct Competition with Scar/WAVE Complex for Rac1.
Structure, 29, 2021
|
|
7B68
 
 | | Crystal structure of MurE from E.coli in complex with Z57299526 | | Descriptor: | 4-[(4-methylphenyl)methyl]-1,4-thiazinane 1,1-dioxide, DIMETHYL SULFOXIDE, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
5IY6
 
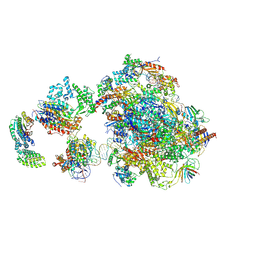 | | Human holo-PIC in the closed state | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
7AJU
 
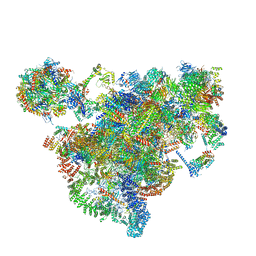 | | Cryo-EM structure of the 90S-exosome super-complex (state Post-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
7AO4
 
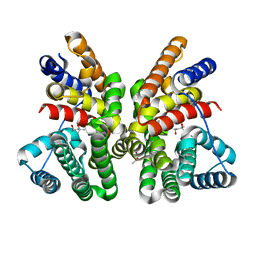 | | Crystal structure of CotB2 variant W288G | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclooctat-9-en-7-ol synthase | | Authors: | Dimos, N, Driller, R, Loll, B. | | Deposit date: | 2020-10-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Impression of a Nonexisting Catalytic Effect: The Role of CotB2 in Guiding the Complex Biosynthesis of Cyclooctat-9-en-7-ol.
J.Am.Chem.Soc., 142, 2020
|
|
1FS9
 
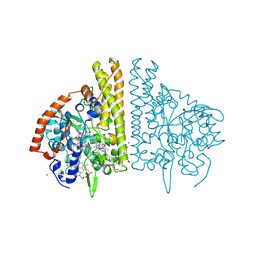 | | CYTOCHROME C NITRITE REDUCTASE FROM WOLINELLA SUCCINOGENES-AZIDE COMPLEX | | Descriptor: | AZIDE ION, CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, ... | | Authors: | Einsle, O, Stach, P, Messerschmidt, A, Simon, J, Kroeger, A, Huber, R, Kroneck, P.M.H. | | Deposit date: | 2000-09-08 | | Release date: | 2001-01-17 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 A resolution, inhibitor binding, and heme-packing motifs.
J.Biol.Chem., 275, 2000
|
|
4W99
 
 | | Apo-structure of the Y79F,W322E-double mutant of Etr1p | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH, B-specific] 1, mitochondrial, ... | | Authors: | Quade, N, Voegeli, B, Rosenthal, R, Erb, T.J. | | Deposit date: | 2014-08-27 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The use of ene adducts to study and engineer enoyl-thioester reductases.
Nat.Chem.Biol., 11, 2015
|
|
