3W9J
 
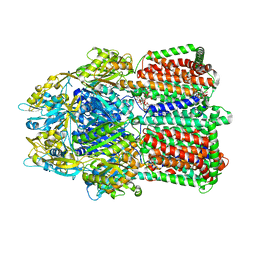 | | Structural basis for the inhibition of bacterial multidrug exporters | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, Multidrug resistance protein MexB, [{2-[({[(3R)-1-{8-[(4-tert-butyl-1,3-thiazol-2-yl)carbamoyl]-4-oxo-3-[(E)-2-(1H-tetrazol-5-yl)ethenyl]-4H-pyrido[1,2-a]pyrimidin-2-yl}piperidin-3-yl]oxy}carbonyl)amino]ethyl}(dimethyl)ammonio]acetate | | 著者 | Sakurai, K, Nakashima, R, Hayashi, K, Yamaguchi, A. | | 登録日 | 2013-04-04 | | 公開日 | 2013-07-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structural basis for the inhibition of bacterial multidrug exporters
Nature, 500, 2013
|
|
3WBK
 
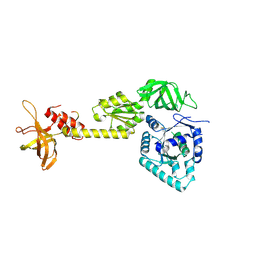 | | crystal structure analysis of eukaryotic translation initiation factor 5B and 1A complex | | 分子名称: | Eukaryotic translation initiation factor 1A, Eukaryotic translation initiation factor 5B | | 著者 | Zheng, A, Yamamoto, R, Ose, T, Yu, J, Tanaka, I, Yao, M. | | 登録日 | 2013-05-20 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | X-ray structures of eIF5B and the eIF5B-eIF1A complex: the conformational flexibility of eIF5B is restricted on the ribosome by interaction with eIF1A
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WAL
 
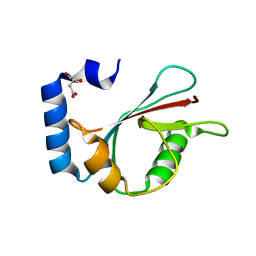 | | Crystal structure of human LC3A_2-121 | | 分子名称: | D-MALATE, Microtubule-associated proteins 1A/1B light chain 3A | | 著者 | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | 登録日 | 2013-05-06 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3W2G
 
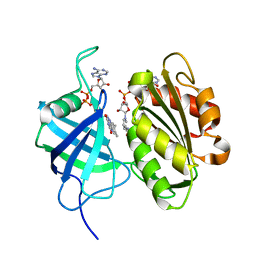 | | Crystal structure of fully reduced form of NADH-cytochrome b5 reductase from pig liver | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | 登録日 | 2012-11-28 | | 公開日 | 2013-07-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
6IRX
 
 | |
3W2F
 
 | | Crystal structure of oxidation intermediate (10 min) of NADH-cytochrome b5 reductase from pig liver | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | 登録日 | 2012-11-28 | | 公開日 | 2013-07-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
4MOV
 
 | | 1.45 A Resolution Crystal Structure of Protein Phosphatase 1 | | 分子名称: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | 著者 | Choy, M.S, Peti, W, Page, R. | | 登録日 | 2013-09-12 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.4503 Å) | | 主引用文献 | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4E4J
 
 | | Crystal structure of arginine deiminase from Mycoplasma penetrans | | 分子名称: | Arginine deiminase, CHLORIDE ION | | 著者 | Benach, J, Gallego, P, Planell, R, Querol, E, Perez Pons, J.A, Reverter, D. | | 登録日 | 2012-03-13 | | 公開日 | 2012-10-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Characterization of the Enzymes Composing the Arginine Deiminase Pathway in Mycoplasma penetrans.
Plos One, 7, 2012
|
|
6IRY
 
 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH | | 分子名称: | 1,2-ETHANEDIOL, PDX1 C-terminal-inhibiting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | 登録日 | 2018-11-14 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
7DU4
 
 | |
7DU5
 
 | |
3WBH
 
 | | Structural characteristics of alkaline phosphatase from a moderately halophilic bacteria Halomonas sp.593 | | 分子名称: | Alkaline phosphatase, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Arai, S, Yonezawa, Y, Ishibashi, M, Matsumoto, F, Tamada, T, Tokunaga, H, Tokunaga, M, Kuroki, R. | | 登録日 | 2013-05-17 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural characteristics of alkaline phosphatase from the moderately halophilic bacterium Halomonas sp. 593.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WG1
 
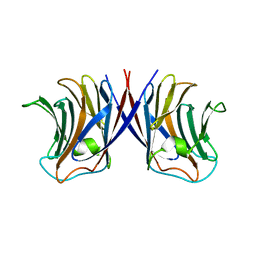 | | Crystal structure of Agrocybe cylindracea galectin with lactose | | 分子名称: | Galactoside-binding lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Kuwabara, N, Hu, D, Tateno, H, Makio, H, Hirabayashi, J, Kato, R. | | 登録日 | 2013-07-25 | | 公開日 | 2013-11-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Conformational change of a unique sequence in a fungal galectin from Agrocybe cylindracea controls glycan ligand-binding specificity.
Febs Lett., 587, 2013
|
|
4LHF
 
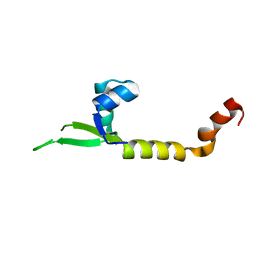 | | Crystal structure of a DNA binding protein from phage P2 | | 分子名称: | Regulatory protein cox | | 著者 | Berntsson, R.P.-A, Odegrip, R, Sehlen, W, Skaar, K, Svensson, L.M, Massad, T, Haggard-Ljungquist, E, Stenmark, P. | | 登録日 | 2013-07-01 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | Structural insight into DNA binding and oligomerization of the multifunctional Cox protein of bacteriophage P2.
Nucleic Acids Res., 42, 2014
|
|
6I9B
 
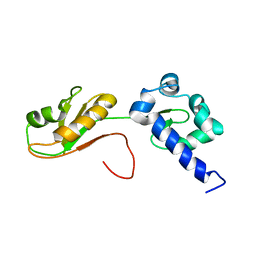 | | NMR structure of the La module from human LARP4A | | 分子名称: | La-related protein 4 | | 著者 | Conte, M.R, Martino, L, Atkinson, R.A, Kelly, G, Cruz-Gallardo, I, De Tito, S, Trotta, R. | | 登録日 | 2018-11-22 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | LARP4A recognizes polyA RNA via a novel binding mechanism mediated by disordered regions and involving the PAM2w motif, revealing interplay between PABP, LARP4A and mRNA.
Nucleic Acids Res., 47, 2019
|
|
7E4S
 
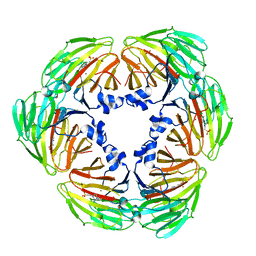 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with HEPES | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-dehydro-4-deoxy-D-glucuronate isomerase, ZINC ION | | 著者 | Yamamoto, Y, Takase, R, Mikami, B, Hashimoto, W. | | 登録日 | 2021-02-15 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Crystal structures of Lacticaseibacillus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI in complex with substrate analogs
J.Appl.Glyosci., 2023
|
|
1R2T
 
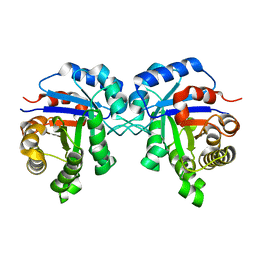 | |
3W2H
 
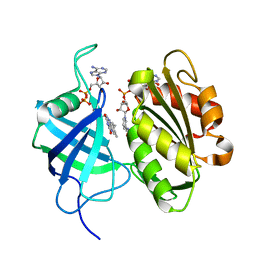 | | Crystal structure of oxidation intermediate (1min) of NADH-cytochrome b5 reductase from pig liver | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | 登録日 | 2012-11-28 | | 公開日 | 2013-07-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.752 Å) | | 主引用文献 | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
7E6E
 
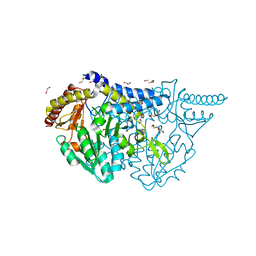 | | Crystal structure of PMP-bound form of cysteine desulfurase SufS R376A from Bacillus subtilis in D-cycloserine-inhibition | | 分子名称: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Cysteine desulfurase SufS, ... | | 著者 | Nakamura, R, Takahashi, Y, Fujishiro, T. | | 登録日 | 2021-02-22 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
3W39
 
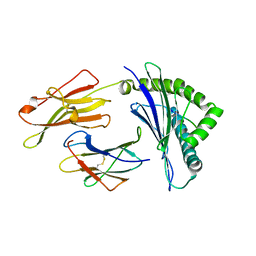 | | Crystal structure of HLA-B*5201 in complexed with HIV immunodominant epitope (TAFTIPSI) | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-52 alpha chain, ... | | 著者 | Yagita, Y, Kuse, N, Kuroki, K, Gatanaga, H, Carlson, J.M, Chikata, T, Brumme, Z.L, Murakoshi, H, Akahoshi, T, Pfeifer, N, Mallal, S, John, M, Ose, T, Matsubara, H, Kanda, R, Fukunaga, Y, Honda, K, Kawashima, Y, Ariumi, Y, Oka, S, Maenaka, K, Takiguchi, M. | | 登録日 | 2012-12-13 | | 公開日 | 2013-02-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Distinct HIV-1 Escape Patterns Selected by Cytotoxic T Cells with Identical Epitope Specificity
J.Virol., 87, 2013
|
|
7E6A
 
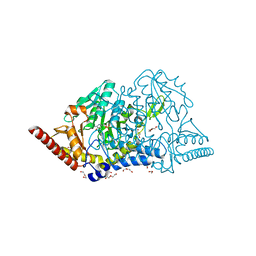 | | Crystal structure of cysteine desulfurase SufS C361A from Bacillus subtilis | | 分子名称: | 1,2-ETHANEDIOL, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Nakamura, R, Takahashi, Y, Fujishiro, T. | | 登録日 | 2021-02-22 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
6JBO
 
 | | Crystal structure of EfeO-like protein Algp7 containing samarium ion | | 分子名称: | 1,2-ETHANEDIOL, Alginate-binding protein, CITRIC ACID | | 著者 | Okumura, K, Takase, R, Maruyama, Y, Mikami, B, Murata, K, Hashimoto, W. | | 登録日 | 2019-01-26 | | 公開日 | 2020-01-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Rare metal binding by a cell-surface component of bacterial EfeUOB iron importer
To Be Published
|
|
6JCS
 
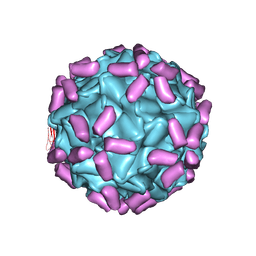 | | AAV5 in complex with AAVR | | 分子名称: | Capsid protein, Dyslexia-associated protein KIAA0319-like protein | | 著者 | Lou, Z, Zhang, R. | | 登録日 | 2019-01-30 | | 公開日 | 2019-08-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
7E6C
 
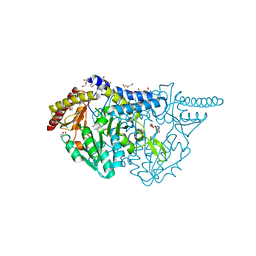 | | Crystal structure of L-cycloserine-bound form of cysteine desulfurase SufS C361A from Bacillus subtilis | | 分子名称: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, 1,2-ETHANEDIOL, Cysteine desulfurase SufS, ... | | 著者 | Nakamura, R, Takahashi, Y, Fujishiro, T. | | 登録日 | 2021-02-22 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
3W92
 
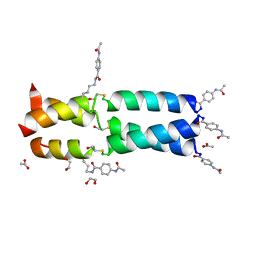 | | Crystal Structure Analysis of the synthetic GCN4 Thioester coiled coil peptide | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, PARA ACETAMIDO BENZOIC ACID, ... | | 著者 | Shahar, A, Zarivach, R, Ashkenasy, G. | | 登録日 | 2013-03-24 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | A high-resolution structure that provides insight into coiled-coil thiodepsipeptide dynamic chemistry
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
