7N5W
 
 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 23) | | 分子名称: | 1,2-ETHANEDIOL, DNA Strand I, DNA Strand II, ... | | 著者 | Horton, J.R, Ren, R, Cheng, X. | | 登録日 | 2021-06-06 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
6YB7
 
 | | SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19). | | 分子名称: | 3C-like proteinase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE | | 著者 | Owen, C.D, Lukacik, P, Strain-Damerell, C.M, Douangamath, A, Powell, A.J, Fearon, D, Brandao-Neto, J, Crawshaw, A.D, Aragao, D, Williams, M, Flaig, R, Hall, D.R, McAuley, K.E, Mazzorana, M, Stuart, D.I, von Delft, F, Walsh, M.A. | | 登録日 | 2020-03-16 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | COVID-19 main protease with unliganded active site
To Be Published
|
|
7N5V
 
 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 20) | | 分子名称: | DNA Strand I, DNA Strand II, ZINC ION, ... | | 著者 | Horton, J.R, Ren, R, Cheng, X. | | 登録日 | 2021-06-06 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.08 Å) | | 主引用文献 | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
6Y2G
 
 | | Crystal structure (orthorhombic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | 分子名称: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | 著者 | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | 登録日 | 2020-02-15 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
8JOP
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with 11a | | 分子名称: | 3C-like proteinase nsp5, methyl (6~{R})-5-ethanoyl-7-oxidanylidene-6-[4-(trifluoromethyl)phenyl]-8,9,10,11-tetrahydro-6~{H}-benzo[b][1,4]benzodiazepine-2-carboxylate | | 著者 | Zeng, R, Liu, Y.Z, Wang, F.L, Yang, S.Y, Lei, J. | | 登録日 | 2023-06-08 | | 公開日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery of benzodiazepine derivatives as a new class of covalent inhibitors of SARS-CoV-2 main protease.
Bioorg.Med.Chem.Lett., 92, 2023
|
|
6RV3
 
 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 1000493 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | 著者 | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | 登録日 | 2019-05-30 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
8J12
 
 | | Cryo-EM structure of the AsCas12f-sgRNA-target DNA ternary complex | | 分子名称: | DNA (38-MER), MAGNESIUM ION, RNA (247-MER), ... | | 著者 | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | 登録日 | 2023-04-12 | | 公開日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Minimal and most efficient genome editing Cas enzyme
To Be Published
|
|
5EYN
 
 | | Crystal structure of Fructokinase from Vibrio cholerae O395 in fructose, ADP, Beryllium trifluoride and calcium ion bound form | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | 著者 | Paul, R, Nath, S, Sen, U. | | 登録日 | 2015-11-25 | | 公開日 | 2016-11-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.289 Å) | | 主引用文献 | Crystal structure of Fructokinase from Vibrio cholerae O395 in apo form
To Be Published
|
|
6YI9
 
 | | Crystal structure of the rat cytosolic PCK1, acetylated on Lys244 | | 分子名称: | 1,2-ETHANEDIOL, Phosphoenolpyruvate carboxykinase, cytosolic [GTP] | | 著者 | Latorre-Muro, P, Baeza, J, Hurtado-Guerrero, R, Hicks, T, Delso, I, Hernandez-Ruiz, C, Velazquez-Campoy, A, Lawton, A.J, Angulo, J, Denu, J.M, Carrodeguas, J.A. | | 登録日 | 2020-04-01 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Self-acetylation at the active site of phosphoenolpyruvate carboxykinase (PCK1) controls enzyme activity.
J.Biol.Chem., 296, 2021
|
|
7N8W
 
 | |
7N8V
 
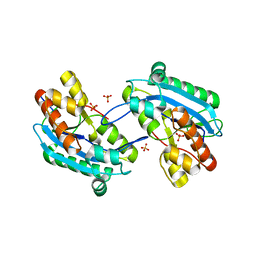 | |
3MMZ
 
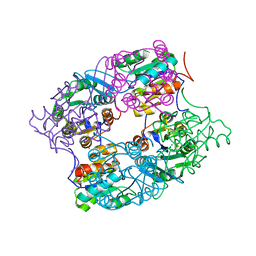 | | CRYSTAL STRUCTURE OF putative HAD family hydrolase from Streptomyces avermitilis MA-4680 | | 分子名称: | CALCIUM ION, CHLORIDE ION, putative HAD family hydrolase | | 著者 | Malashkevich, V.N, Ramagopal, U.A, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-04-20 | | 公開日 | 2010-04-28 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
6YLH
 
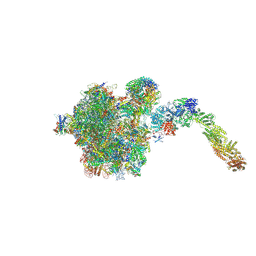 | |
6YLT
 
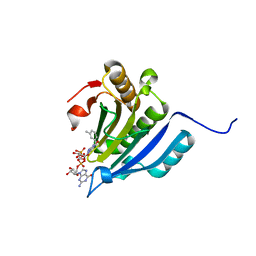 | | Translation initiation factor 4E in complex with 3-MeBn7GpppG mRNA 5' cap analog | | 分子名称: | Eukaryotic translation initiation factor 4E, [[(2~{R},3~{S},4~{R},5~{R})-5-[2-azanyl-7-[(3-methylphenyl)methyl]-6-oxidanylidene-1~{H}-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] hydrogen phosphate | | 著者 | Kubacka, D, Wojcik, R, Baranowski, M.R, Kowalska, J, Jemielity, J. | | 登録日 | 2020-04-07 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Novel N7-Arylmethyl Substituted Dinucleotide mRNA 5' cap Analogs: Synthesis and Evaluation as Modulators of Translation.
Pharmaceutics, 13, 2021
|
|
6Y2F
 
 | | Crystal structure (monoclinic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | 著者 | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | 登録日 | 2020-02-15 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
8J1J
 
 | | Cryo-EM structure of the AsCas12f-YHAM-sgRNAS3-5v7-target DNA | | 分子名称: | DNA (38-MER), MAGNESIUM ION, RNA (118-MER), ... | | 著者 | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | 登録日 | 2023-04-13 | | 公開日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Minimal and most efficient genome editing Cas enzyme
To Be Published
|
|
6Y3C
 
 | | Human COX-1 Crystal Structure | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, Prostaglandin G/H synthase 1, ... | | 著者 | Miciaccia, M, Belviso, B.D, Iaselli, M, Ferorelli, S, Perrone, M.G, Caliandro, R, Scilimati, A. | | 登録日 | 2020-02-18 | | 公開日 | 2020-02-26 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.361 Å) | | 主引用文献 | Three-dimensional structure of human cyclooxygenase (hCOX)-1.
Sci Rep, 11, 2021
|
|
8J3R
 
 | | Cryo-EM structure of the AsCas12f-HKRA-sgRNAS3-5v7-target DNA | | 分子名称: | DNA (37-MER), DNA (38-MER), MAGNESIUM ION, ... | | 著者 | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | 登録日 | 2023-04-18 | | 公開日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Minimal and most efficient genome editing Cas enzyme
To Be Published
|
|
2ZIN
 
 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with BocLys and an ATP analogue | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, N~6~-(tert-butoxycarbonyl)-L-lysine, ... | | 著者 | Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2008-02-19 | | 公開日 | 2008-12-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Multistep Engineering of Pyrrolysyl-tRNA Synthetase to Genetically Encode N(varepsilon)-(o-Azidobenzyloxycarbonyl) lysine for Site-Specific Protein Modification
Chem.Biol., 15, 2008
|
|
8JHE
 
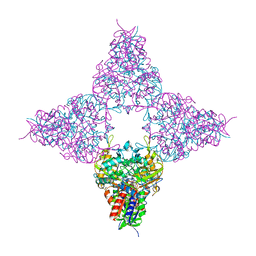 | | Hyper-thermostable ancestral L-amino acid oxidase 2 (HTAncLAAO2) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Hyper thermostable ancestral L-amino acid oxidase | | 著者 | Kawamura, Y, Ishida, C, Miyata, R, Miyata, A, Hayashi, S, Fujinami, D, Ito, S, Nakano, S. | | 登録日 | 2023-05-23 | | 公開日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.201 Å) | | 主引用文献 | Structural and functional analysis of hyper-thermostable ancestral L-amino acid oxidase that can convert Trp derivatives to D-forms by chemoenzymatic reaction.
Commun Chem, 6, 2023
|
|
6Y4C
 
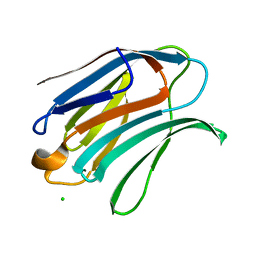 | | Structure of galectin-3C in complex with lactose determined by serial crystallography using an XtalTool support | | 分子名称: | CHLORIDE ION, Galectin-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Shilova, A, Hakansson, M, Welin, M, Kovacic, R, Mueller, U, Logan, D.T. | | 登録日 | 2020-02-20 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Current status and future opportunities for serial crystallography at MAX IV Laboratory.
J.Synchrotron Radiat., 27, 2020
|
|
2ZCE
 
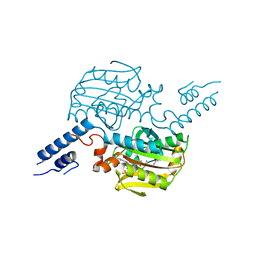 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with pyrrolysine and an ATP analogue | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PYRROLYSINE, ... | | 著者 | Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-11-08 | | 公開日 | 2008-04-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystallographic Studies on Multiple Conformational States of Active-site Loops in Pyrrolysyl-tRNA Synthetase
J.Mol.Biol., 378, 2008
|
|
6Y9E
 
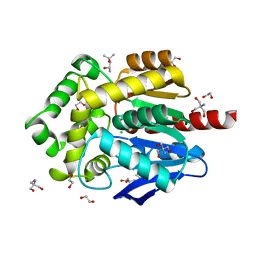 | | Crystal structure of putative ancestral haloalkane dehalogenase AncHLD2 (node 2) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Chaloupkova, R, Damborsky, J, Marek, M. | | 登録日 | 2020-03-09 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structures of hyperstable ancestral haloalkane dehalogenases show restricted conformational dynamics.
Comput Struct Biotechnol J, 18, 2020
|
|
6RT4
 
 | |
7NKV
 
 | |
