6X2Q
 
 | | Complex of Gynuella sunshinyii GH46 chitosanase GsCsn46A with chitotetraose | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Chitosanase | | Authors: | Qin, Z, Wang, Y. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Complex of Gynuella sunshinyii GH46 chitosanase GsCsn46A with chitotetraose
To Be Published
|
|
4WTR
 
 | | Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaribiose | | Descriptor: | beta-1,3-glucanosyltransferase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Qin, Z, Yan, Q, Lei, J, Yang, S, Jiang, Z. | | Deposit date: | 2014-10-30 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The first crystal structure of a glycoside hydrolase family 17 beta-1,3-glucanosyltransferase displays a unique catalytic cleft.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WTP
 
 | | Crystal structure of glycoside hydrolase family 17 beta-1,3-glucanosyltransferase from Rhizomucor miehei | | Descriptor: | 1,2-ETHANEDIOL, beta-1,3-glucanosyltransferase | | Authors: | Qin, Z, Yan, Q, Lei, J, Yang, S, Jiang, Z. | | Deposit date: | 2014-10-30 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The first crystal structure of a glycoside hydrolase family 17 beta-1,3-glucanosyltransferase displays a unique catalytic cleft.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WTS
 
 | | Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaritriose | | Descriptor: | beta-1,3-glucanosyltransferase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Qin, Z, Yan, Q, Lei, J, Yang, S, Jiang, Z. | | Deposit date: | 2014-10-30 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The first crystal structure of a glycoside hydrolase family 17 beta-1,3-glucanosyltransferase displays a unique catalytic cleft.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WY5
 
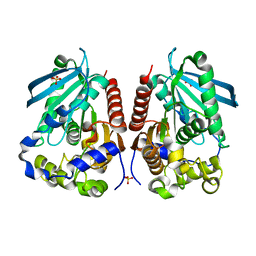 | | Structural analysis of two fungal esterases from Rhizomucor miehei explaining their substrate specificity | | Descriptor: | Esterase, SULFATE ION | | Authors: | Qin, Z, Yang, S, Duan, X, Yan, Q, Jiang, Z. | | Deposit date: | 2014-11-15 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural insights into the substrate specificity of two esterases from the thermophilic Rhizomucor miehei
J.Lipid Res., 56, 2015
|
|
4ZM6
 
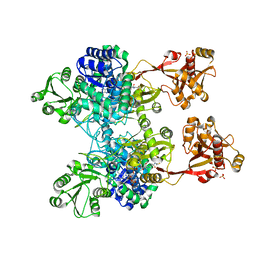 | | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase | | Descriptor: | ACETYL COENZYME *A, N-acetyl-beta-D glucosaminidase, SULFATE ION | | Authors: | Qin, Z, Xiao, Y, Yang, X, Jiang, Z, Yang, S, Mesters, J.R. | | Deposit date: | 2015-05-02 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain glycoside hydrolase family 3 beta-N-acetylglucosaminidase
Sci Rep, 5, 2015
|
|
4WY8
 
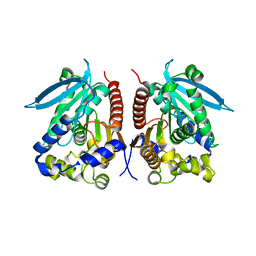 | | Structural analysis of two fungal esterases from Rhizomucor miehei explaining their substrate specificity | | Descriptor: | esterase | | Authors: | Qin, Z, Yang, S, Duan, X, Yan, Q, Jiang, Z. | | Deposit date: | 2014-11-16 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural insights into the substrate specificity of two esterases from the thermophilic Rhizomucor miehei
J.Lipid Res., 56, 2015
|
|
5JVV
 
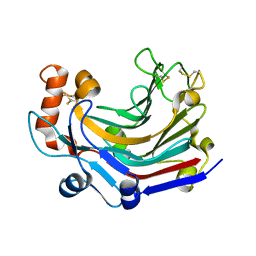 | | Crystal structure and characterization an elongating GH family 16 beta-1,3-glucosyltransferase | | Descriptor: | beta-1,3-glucosyltransferase | | Authors: | Qin, Z, Yan, Q, Yang, S, Jiang, Z. | | Deposit date: | 2016-05-11 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Catalytic Mechanism of a Novel Glycoside Hydrolase Family 16 "Elongating" beta-Transglycosylase
J. Biol. Chem., 292, 2017
|
|
6LWU
 
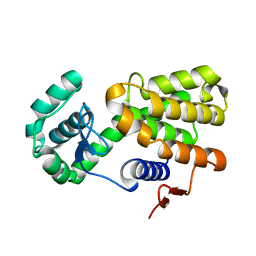 | | Complex of Gynuella sunshinyii GH46 chitosanase GsCsn46A E19A with chitopentaose | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, chitosanase | | Authors: | Qin, Z, Wang, Y. | | Deposit date: | 2020-02-09 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.419 Å) | | Cite: | Complex of Gynuella sunshinyii GH46 chitosanase GsCsn46A E19A with chitopentaose
To Be Published
|
|
5XC2
 
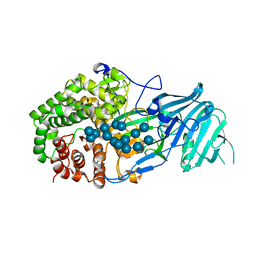 | | Crystal structure of GH family 81 beta-1,3-glucanase from Rhizomucr miehei complexed with laminarihexaose | | Descriptor: | Endo-beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Qin, Z, Yang, S, Peng, Z, Yan, Q, Jiang, Z. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytic mechanism of glycoside hydrolase family 81 beta-1,3-glucanase
To Be Published
|
|
6A6C
 
 | |
5WVP
 
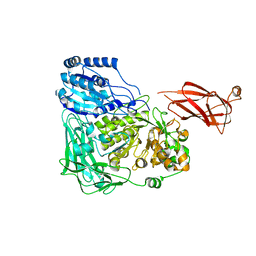 | | Expression, characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii | | Descriptor: | Beta-glucosidase, beta-D-mannopyranose | | Authors: | Jiang, Z, Wu, S, Yang, D, Qin, Z, You, X, Huang, P. | | Deposit date: | 2016-12-28 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Expression, characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii
To Be Published
|
|
8JB4
 
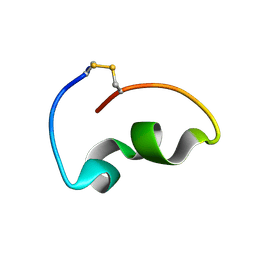 | | lipopolysaccharide-binding domain-LBDB | | Descriptor: | Antilipopolysaccharide factor D | | Authors: | Huang, J, Qin, Z. | | Deposit date: | 2023-05-08 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Machine learning and genetic algorithm-guided directed evolution for the development of small-molecule antibiotics originating from antimicrobial peptides
To Be Published
|
|
5KLV
 
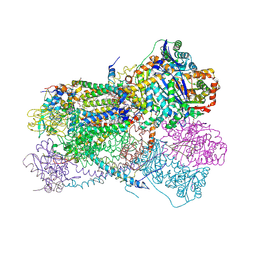 | | Structure of bos taurus cytochrome bc1 with fenamidone inhibited | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl octadecanoate, (5S)-5-methyl-2-(methylsulfanyl)-5-phenyl-3-(phenylamino)-3,5-dihydro-4H-imidazol-4-one, 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Zhou, Y, Xiao, Y, Tang, W.K, Yu, C.A, Qin, Z. | | Deposit date: | 2016-06-25 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Hydrogen Bonding to the Substrate Is Not Required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III.
J.Biol.Chem., 291, 2016
|
|
8C5V
 
 | |
4QP0
 
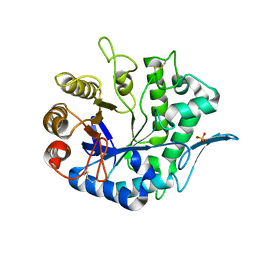 | | Crystal Structure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei | | Descriptor: | Endo-beta-mannanase, SULFATE ION | | Authors: | Zheng, Q.J, Peng, Z, Liu, Y, Yan, Q.J, Chen, Z.Z, Qin, Z. | | Deposit date: | 2014-06-22 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5XBZ
 
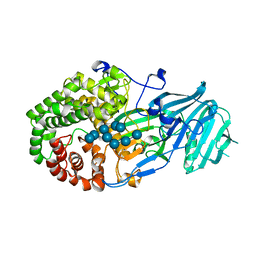 | | Crystal structure of GH family 81 beta-1,3-glucanase from Rhizomucr miehei complexed with laminaripentaose | | Descriptor: | Endo-beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Yang, S, Qin, Z, Zhou, P, Yan, Q, Jiang, Z. | | Deposit date: | 2017-03-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytic mechanism of glycoside hydrolase family 81 beta-1,3-glucanase
To Be Published
|
|
5WUG
 
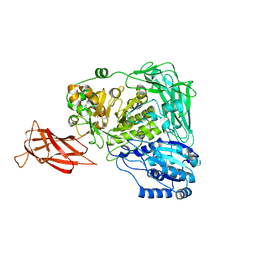 | | Expression, characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii | | Descriptor: | Beta-glucosidase | | Authors: | Jiang, Z, Wu, S, Yang, D, Qin, Z, You, X, Huang, P. | | Deposit date: | 2016-12-17 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.216 Å) | | Cite: | Expression, Biochemical Characterization and Structure Resolution of beta-glucosidase from Paenibacillus barengoltzii
J Food Sci Technol(China), 2019
|
|
6JHI
 
 | | Crystal structure of mutant D470A of Pullulanase from Paenibacillus barengoltzii complexed with maltotetraose | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pulullanase, ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | Crystal structure of mutant D470A of Pullulanase from Paenibacillus barengoltzii complexed with maltotetraose
To Be Published
|
|
6JHH
 
 | | Crystal structure of mutant D350A of Pullulanase from Paenibacillus barengoltzii complexed with maltotriose | | Descriptor: | CALCIUM ION, Pulullanase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.025 Å) | | Cite: | Crystal structure of mutant D350A of Pullulanase from Paenibacillus barengoltzii complexed with maltotriose
To Be Published
|
|
6JHG
 
 | | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii in space group P212121 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pulullanase | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii in space group P212121
To Be Published
|
|
5YUQ
 
 | | The high resolution structure of chitinase (RmChi1) from the thermophilic fungus Rhizomucor miehei (sp P1) | | Descriptor: | Chintase | | Authors: | Jiang, Z.Q, Hu, S.Q, Liu, Y.C, Qin, Z, Yan, Q.J, Yang, S.Q. | | Deposit date: | 2017-11-23 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of a chitinase (RmChiA) from the thermophilic fungus Rhizomucor miehei with a real active site tunnel.
Biochim Biophys Acta Proteins Proteom, 2021
|
|
5XWF
 
 | | Crystal structure of chitinase (RmChi1) from Rhizomucor miehei (SP3221/SAD) | | Descriptor: | Fungal chitinase from Rhizomucor miehei (SeMet-substituted proteins) | | Authors: | Jiang, Z.Q, Hu, S.Q, Liu, Y.C, Qin, Z, Yan, Q.J, Yang, S.Q. | | Deposit date: | 2017-06-29 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.581 Å) | | Cite: | Crystal structure of a chitinase (RmChiA) from the thermophilic fungus Rhizomucor miehei with a real active site tunnel.
Biochim Biophys Acta Proteins Proteom, 2021
|
|
6JFX
 
 | | Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with maltopentaose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-12 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6JHF
 
 | | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
