2OP7
 
 | | WW4 | | Descriptor: | NEDD4-like E3 ubiquitin-protein ligase WWP1 | | Authors: | Qin, H.N, Li, M.F, Pu, H, Sankaran, S, Ahmed, S, Song, J.X. | | Deposit date: | 2007-01-27 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the forth WW domain of WWP1
To be Published
|
|
3GXU
 
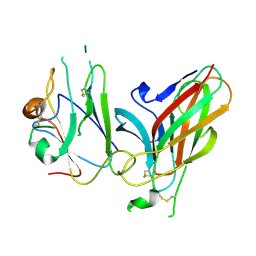 | | Crystal structure of Eph receptor and ephrin complex | | Descriptor: | Ephrin type-A receptor 4, Ephrin-B2 | | Authors: | Qin, H.N, Song, J.X. | | Deposit date: | 2009-04-03 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural characterization of the EphA4-ephrin-B2 complex reveals new features enabling Eph-ephrin binding promiscuity
J.Biol.Chem., 2009
|
|
3WLX
 
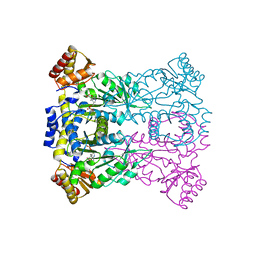 | | Crystal structure of low-specificity L-threonine aldolase from Escherichia coli | | Descriptor: | Low specificity L-threonine aldolase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Qin, H.-M, Imai, F.L, Miyakawa, T, Kataoka, M, Okai, M, Hou, F, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2013-11-15 | | Release date: | 2014-12-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure analysis of L-threonine aldolase from Escherichia coli unravels the low-specificity and thermostability
To be Published
|
|
3YGS
 
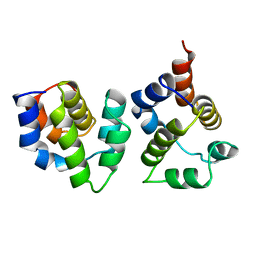 | | APAF-1 CARD IN COMPLEX WITH PRODOMAIN OF PROCASPASE-9 | | Descriptor: | APOPTOTIC PROTEASE ACTIVATING FACTOR 1, PROCASPASE 9 | | Authors: | Qin, H, Srinivasula, S, Wu, G, Fernandes-Alnemri, T, Alnemri, E, Shi, Y. | | Deposit date: | 1999-05-08 | | Release date: | 2000-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1.
Nature, 399, 1999
|
|
4H8N
 
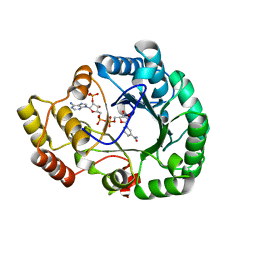 | | Crystal structure of conjugated polyketone reductase C2 from candida parapsilosis complexed with NADPH | | Descriptor: | Conjugated polyketone reductase C2, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Qin, H.-M, Yamamura, A, Miyakawa, T, Maruoka, S, Ohtsuka, J, Nagata, K, Kataoka, M, Shimizu, S, Tanokura, M. | | Deposit date: | 2012-09-23 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of conjugated polyketone reductase from Candida parapsilosis IFO 0708 reveals conformational changes for substrate recognition upon NADPH binding
Appl.Microbiol.Biotechnol., 98, 2014
|
|
5GMT
 
 | |
3WG6
 
 | | Crystal structure of conjugated polyketone reductase C1 from Candida parapsilosis complexed with NADPH | | Descriptor: | Conjugated polyketone reductase C1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Qin, H.-M, Yamamura, A, Miyakawa, T, Maruoka, S, Ohtsuka, J, Nagata, K, Kataoka, M, Shimizu, S, Tanokura, M. | | Deposit date: | 2013-07-28 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of conjugated polyketone reductase (CPR-C1) from Candida parapsilosis IFO 0708 complexed with NADPH.
Proteins, 81, 2013
|
|
2LW8
 
 | |
2N6E
 
 | |
5WUT
 
 | |
5Y33
 
 | |
3WGC
 
 | | Aeromonas jandaei L-allo-threonine aldolase H128Y/S292R double mutant | | Descriptor: | L-allo-threonine aldolase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Qin, H.M, Imai, F.L, Miyakawa, T, Kataoka, M, Okai, M, Ohtsuka, J, Hou, F, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2013-08-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | L-allo-Threonine aldolase with an H128Y/S292R mutation from Aeromonas jandaei DK-39 reveals the structural basis of changes in substrate stereoselectivity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WGB
 
 | | Crystal structure of aeromonas jandaei L-allo-threonine aldolase | | Descriptor: | GLYCINE, L-allo-threonine aldolase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Qin, H.M, Imai, F.L, Miyakawa, T, Kataoka, M, Okai, M, Ohtsuka, J, Hou, F, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2013-08-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | L-allo-Threonine aldolase with an H128Y/S292R mutation from Aeromonas jandaei DK-39 reveals the structural basis of changes in substrate stereoselectivity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3W20
 
 | | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase from Burkholderia ambifaria AMMD | | Descriptor: | Putative uncharacterized protein, ZINC ION | | Authors: | Qin, H.M, Miyakawa, T, Jia, M.Z, Nakamura, A, Ohtsuka, J, Xue, Y.L, Kawashima, T, Kasahara, T, Hibi, M, Ogawa, J, Tanokura, M. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase from Burkholderia ambifaria AMMD
Plos One, 8, 2013
|
|
3VXG
 
 | | Crystal structure of conjugated polyketone reductase C2 from Candida Parapsilosis | | Descriptor: | Conjugated polyketone reductase C2 | | Authors: | Qin, H.-M, Yamamura, A, Miyakawa, T, Maruoka, S, Ohtsuka, J, Nagata, K, Kataoka, M, Shimizu, S, Tanokura, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of conjugated polyketone reductase from Candida parapsilosis IFO 0708 reveals conformational changes for substrate recognition upon NADPH binding
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3W21
 
 | | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase in complex with alpha-KG from Burkholderia ambifaria AMMD | | Descriptor: | 2-OXOGLUTARIC ACID, Putative uncharacterized protein, ZINC ION | | Authors: | Qin, H.M, Miyakawa, T, Jia, M.Z, Nakamura, A, Ohtsuka, J, Xue, Y.L, Kawashima, T, Kasahara, T, Hibi, M, Ogawa, J, Tanokura, M. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase from Burkholderia ambifaria AMMD
Plos One, 8, 2013
|
|
2MDK
 
 | |
6JQ9
 
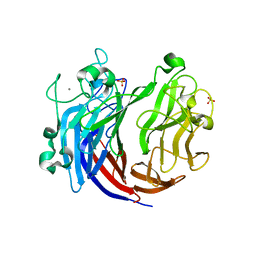 | | Crystal structure of a lyase from Alteromonas sp. | | Descriptor: | CALCIUM ION, SULFATE ION, Short ulvan lyase | | Authors: | Qin, H.M, Guo, Q.Q. | | Deposit date: | 2019-03-29 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Biochemical characterization and structural analysis of ulvan lyase from marine Alteromonas sp. reveals the basis for its salt tolerance.
Int.J.Biol.Macromol., 147, 2020
|
|
2KOU
 
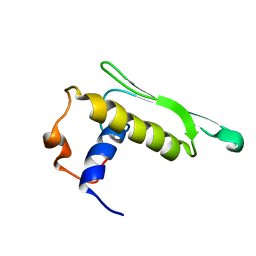 | | DICER LIKE protein | | Descriptor: | Dicer-like protein 4 | | Authors: | Qin, H, Song, J, Yuan, Y.A. | | Deposit date: | 2009-09-30 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Arabidopsis thaliana DCL4 DUF283 domain reveals a noncanonical double-stranded RNA-binding fold for protein-protein interaction
Rna, 2010
|
|
6JQX
 
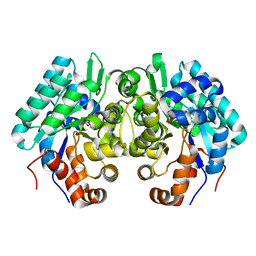 | |
5ZSY
 
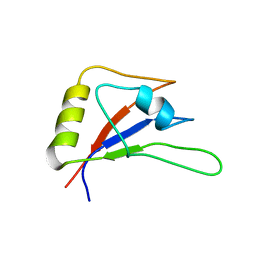 | | RBM10-RRM2 domain and its lung cancer related mutant | | Descriptor: | RNA-binding protein 10 | | Authors: | Qin, H. | | Deposit date: | 2018-04-30 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural, dynamics, and RNA binding comparison between RBM10-RRM2 domain and its lung cancer related mutation.
To be published
|
|
5ZSW
 
 | | RBM10-RRM2 domain and its lung cancer related mutant | | Descriptor: | RNA-binding protein 10 | | Authors: | Qin, H. | | Deposit date: | 2018-04-30 | | Release date: | 2019-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural, dynamics, and RNA binding comparison between RBM10-RRM2 domain and its lung cancer related mutation.
To be published
|
|
6JQW
 
 | |
8DHT
 
 | | Crystal structure of a typeIII Rubisco | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ACETATE ION, GLYCEROL, ... | | Authors: | Qingqiu, H. | | Deposit date: | 2022-06-28 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of a type III Rubisco in complex with its product 3-phosphoglycerate.
Proteins, 91, 2023
|
|
8U1T
 
 | | SARS-CoV-2 Envelope Protein Transmembrane Domain: Dimeric Structure Determined by Solid-State NMR | | Descriptor: | Envelope small membrane protein | | Authors: | Zhang, R, Qin, H, Prasad, R, Fu, R, Zhou, H.X, Cross, T. | | Deposit date: | 2023-09-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Dimeric Transmembrane Structure of the SARS-CoV-2 E Protein.
Commun Biol, 6, 2023
|
|
