8K8H
 
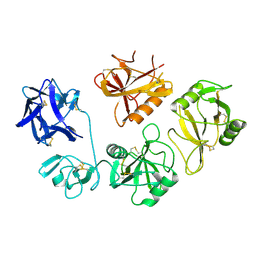 | |
4M5E
 
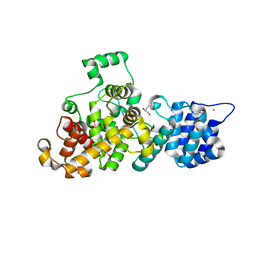 | | Tse3 structure | | Descriptor: | CADMIUM ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Qian, Y. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
7WY5
 
 | | ADGRL3/Gq complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling.
Mol.Cell, 82, 2022
|
|
7WYB
 
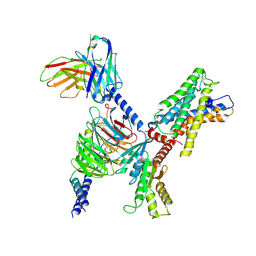 | | ADGRL3/Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and G q , G s , G i , and G 12 coupling.
Mol.Cell, 82, 2022
|
|
7WY8
 
 | | ADGRL3/Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling
Mol.Cell, 82, 2022
|
|
7X10
 
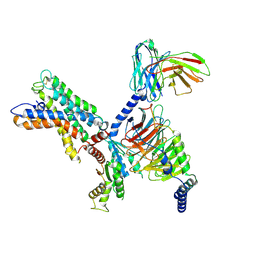 | | ADGRL3/miniG12 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-22 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling.
Mol.Cell, 82, 2022
|
|
4GQB
 
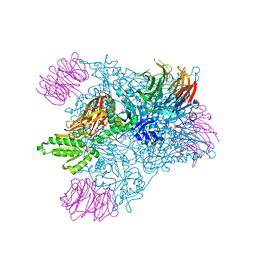 | | Crystal Structure of the human PRMT5:MEP50 Complex | | Descriptor: | (2S,5S,6E)-2,5-diamino-6-[(3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxydihydrofuran-2(3H)-ylidene]hexanoic acid, Histone H4 peptide, Methylosome protein 50, ... | | Authors: | Antonysamy, S, Bonday, Z, Campbell, R, Doyle, B, Druzina, Z, Gheyi, T, Han, B, Jungheim, L.N, Qian, Y, Rauch, C, Russell, M, Sauder, J.M, Wasserman, S.R, Weichert, K, Willard, F.S, Zhang, A, Emtage, S. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the human PRMT5:MEP50 complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4I5I
 
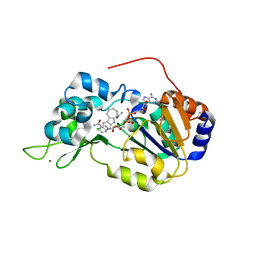 | | Crystal structure of the SIRT1 catalytic domain bound to NAD and an EX527 analog | | Descriptor: | (6S)-2-chloro-5,6,7,8,9,10-hexahydrocyclohepta[b]indole-6-carboxamide, NAD-dependent protein deacetylase sirtuin-1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Zhao, X, Allison, D, Condon, B, Zhang, F, Gheyi, T, Zhang, A, Ashok, S, Russell, M, Macewan, I, Qian, Y, Jamison, J.A, Luz, J.G. | | Deposit date: | 2012-11-28 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 angstrom crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition.
J.Med.Chem., 56, 2013
|
|
1S6C
 
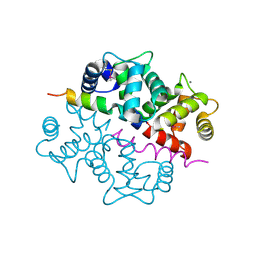 | | Crystal structure of the complex between KChIP1 and Kv4.2 N1-30 | | Descriptor: | CALCIUM ION, Kv4 potassium channel-interacting protein KChIP1b, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhou, W, Qian, Y, Kunjilwar, K, Pfaffinger, P.J, Choe, S. | | Deposit date: | 2004-01-23 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the functional interaction of KChIP1 with Shal-type K(+) channels.
Neuron, 41, 2004
|
|
8WM9
 
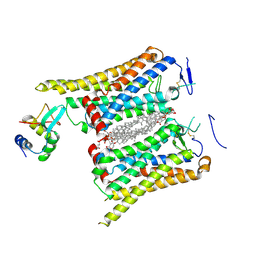 | | Fzd4/DEP complex | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Frizzled-4, Segment polarity protein dishevelled homolog DVL-2 | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis of Frizzled 4 in recognition of Dishevelled 2 unveils mechanism of WNT signaling activation.
Nat Commun, 15, 2024
|
|
8WMA
 
 | | Fzd4/DEP complex (local refined) | | Descriptor: | Frizzled-4, Segment polarity protein dishevelled homolog DVL-2 | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis of Frizzled 4 in recognition of Dishevelled 2 unveils mechanism of WNT signaling activation.
Nat Commun, 15, 2024
|
|
