8OT6
 
 | |
8OTE
 
 | |
7Z23
 
 | | Connexin43 hemi channel in nanodisc | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Qi, C, Korkhov, M.V. | | Deposit date: | 2022-02-25 | | Release date: | 2023-03-08 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structure of the connexin-43 gap junction channel in a putative closed state.
Elife, 12, 2023
|
|
7Z1T
 
 | |
7Z22
 
 | |
7ZXO
 
 | |
7ZXQ
 
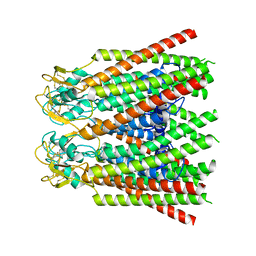 | |
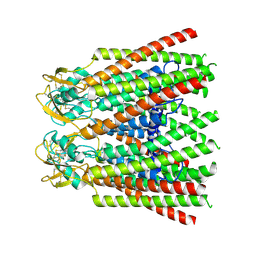
7ZXP
 
 | |
7ZXM
 
 | |
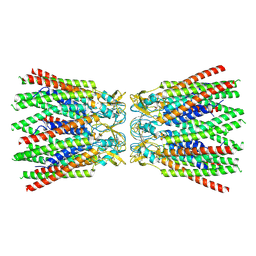
7ZXT
 
 | |
7ZXN
 
 | | cryo-EM structure of Connexin 32 gap junction channel | | Descriptor: | CHOLESTEROL, Gap junction beta-1 protein | | Authors: | Qi, C, Korkhov, V.M. | | Deposit date: | 2022-05-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structures of wild-type and selected CMT1X mutant connexin 32 gap junction channels and hemichannels.
Sci Adv, 9, 2023
|
|
3CEY
 
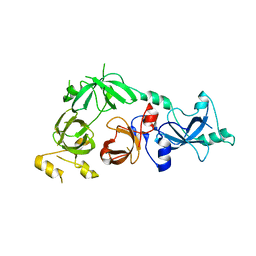 | | Crystal structure of L3MBTL2 | | Descriptor: | Lethal(3)malignant brain tumor-like 2 protein | | Authors: | Nady, N, Guo, Y, Pan, P, Allali-Hassani, A, Qi, C, Zhu, H, Dong, A, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Edwards, A.M, Weigelt, J, Bountra, C, Arrowsmith, C.H, Bochkarev, A, Read, R, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-29 | | Release date: | 2008-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2.
Nucleic Acids Res., 37, 2009
|
|
3F70
 
 | | Crystal structure of L3MBTL2-H4K20me1 complex | | Descriptor: | Lethal(3)malignant brain tumor-like 2 protein, N-METHYL-LYSINE | | Authors: | Guo, Y, Qi, C, Allali-Hassani, A, Pan, P, Zhu, H, Dong, A, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Edwards, A.M, Weigelt, J, Bountra, C, Arrowsmith, C.H, Botchkarev, A, Read, R, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-07 | | Release date: | 2009-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2.
Nucleic Acids Res., 37, 2009
|
|
6R4P
 
 | |
6RMG
 
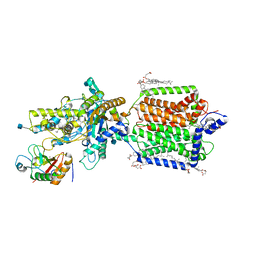 | | Structure of PTCH1 bound to a modified Hedgehog ligand ShhN-C24II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Korkhov, V.M, Qi, C. | | Deposit date: | 2019-05-06 | | Release date: | 2019-10-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of sterol recognition by human hedgehog receptor PTCH1.
Sci Adv, 5, 2019
|
|
6R3Q
 
 | |
5EBD
 
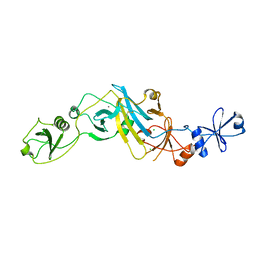 | | Crystal structure of EccB1 of Mycobacterium tuberculosis in spacegroup P21 (state IV) | | Descriptor: | CALCIUM ION, CHLORIDE ION, ESX-1 secretion system protein eccB1 | | Authors: | Zhang, X.L, Qi, C, Xie, X.Q, Li, D.F, Bi, L.J. | | Deposit date: | 2015-10-19 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic observation of the movement of the membrane-distal domain of the T7SS core component EccB1 from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5EBC
 
 | | Crystal structure of EccB1 of Mycobacterium tuberculosis in spacegroup P21 (state III) | | Descriptor: | CALCIUM ION, ESX-1 secretion system protein eccB1 | | Authors: | Zhang, X.L, Qi, C, Xie, X.Q, Li, D.F, Bi, L.J. | | Deposit date: | 2015-10-19 | | Release date: | 2016-02-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic observation of the movement of the membrane-distal domain of the T7SS core component EccB1 from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5YZ2
 
 | | the cystathionine-beta-synthase (CBS) domain of magnesium and cobalt efflux protein CorC in complex with both C2'- and C3'-endo AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Magnesium and cobalt efflux protein CorC | | Authors: | Feng, N, Qi, C, Li, D.F, Wang, D.C. | | Deposit date: | 2017-12-12 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The C2'- and C3'-endo equilibrium for AMP molecules bound in the cystathionine-beta-synthase domain.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
8OTI
 
 | | CTE typeIII tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Tetter, S, Qi, C, Ryskeldi-Falcon, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Tau filaments from amyotrophic lateral sclerosis/parkinsonism-dementia complex adopt the CTE fold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8K11
 
 | | SID1 transmembrane family member 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 2 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K12
 
 | | SID1 transmembrane family member 2 | | Descriptor: | SID1 transmembrane family member 2 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K1B
 
 | | SID1 transmembrane family member 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 1 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K10
 
 | | SID1 transmembrane family member 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 2 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K13
 
 | | SID1 transmembrane family member 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 1 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
