2VN5
 
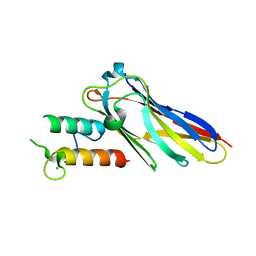 | | The Clostridium cellulolyticum dockerin displays a dual binding mode for its cohesin partner | | Descriptor: | CALCIUM ION, ENDOGLUCANASE A, SCAFFOLDING PROTEIN | | Authors: | Pinheiro, B.A, Prates, J.A.M, Proctor, M.R, Gilbert, H.J, Davies, G.J, Money, V.A, Martinez-Fleites, C, Bayer, E.A, Fontes, C.M.G.A, Fierobe, H.P. | | Deposit date: | 2008-01-31 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Clostridium Cellulolyticum Dockerin Displays a Dual Binding Mode for its Cohesin Partner.
J.Biol.Chem., 283, 2008
|
|
2VN6
 
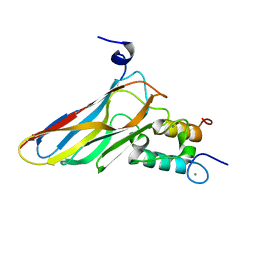 | | The Clostridium cellulolyticum dockerin displays a dual binding mode for its cohesin partner | | Descriptor: | CALCIUM ION, ENDOGLUCANASE A, SCAFFOLDING PROTEIN | | Authors: | Pinheiro, B.A, Prates, J.A.M, Proctor, M.R, Gilbert, H.J, Davies, G.J, Money, V.A, Martinez-Fleites, C, Bayer, E.A, Fontes, C.M.G.A, Fierobe, H.P. | | Deposit date: | 2008-01-31 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The Clostridium Cellulolyticum Dockerin Displays a Dual Binding Mode for its Cohesin Partner.
J.Biol.Chem., 283, 2008
|
|
2BM3
 
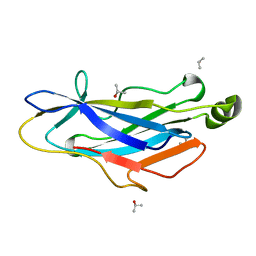 | | Structure of the Type II cohesin from Clostridium thermocellum SdbA | | Descriptor: | ISOPROPYL ALCOHOL, SCAFFOLDING DOCKERIN BINDING PROTEIN A | | Authors: | Carvalho, A.L, Gloster, T.M, Pires, V.M.R, Proctor, M.R, Prates, J.A.M, Ferreira, L.M.A, Turkenburg, J.P, Romao, M.J, Davies, G.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-03-09 | | Release date: | 2005-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights Into the Structural Determinants of Cohesin-Dockerin Specificity Revealed by the Crystal Structure of the Type II Cohesin from Clostridium Thermocellum Sdba.
J.Mol.Biol., 349, 2005
|
|
2IYA
 
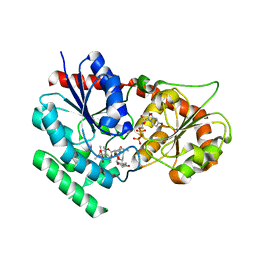 | | The crystal structure of macrolide glycosyltransferases: A blueprint for antibiotic engineering | | Descriptor: | (3S,5R,6S,7R,8R,11R,12S,13R,14S,15S)-6-HYDROXY-5,7,8,11,13,15-HEXAMETHYL-4,10-DIOXO-14-{[3,4,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-XYLO-HEXOPYRANOSYL]OXY}-1,9-DIOXASPIRO[2.13]HEXADEC-12-YL 2,6-DIDEOXY-3-O-METHYL-ALPHA-L-ARABINO-HEXOPYRANOSIDE, OLEANDOMYCIN GLYCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Bolam, D.N, Roberts, S.M, Proctor, M.R, Turkenburg, J.P, Dodson, E.J, Martinez-Fleites, C, Yang, M, Davis, B.G, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2006-07-13 | | Release date: | 2007-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Two Macrolide Glycosyltransferases Provides a Blueprint for Host Cell Antibiotic Immunity.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2IYF
 
 | | The crystal structure of macrolide glycosyltransferases: A blueprint for antibiotic engineering | | Descriptor: | ERYTHROMYCIN A, MAGNESIUM ION, OLEANDOMYCIN GLYCOSYLTRANSFERASE, ... | | Authors: | Bolam, D.N, Roberts, S.M, Proctor, M.R, Turkenburg, J.P, Dodson, E.J, Martinez-Fleites, C, Yang, M, Davis, B.G, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2006-07-17 | | Release date: | 2007-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Two Macrolide Glycosyltransferases Provides a Blueprint for Host Cell Antibiotic Immunity.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
