1IHM
 
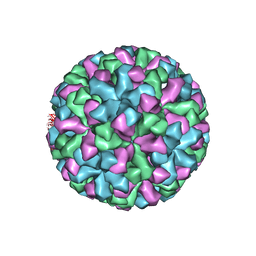 | | CRYSTAL STRUCTURE ANALYSIS OF NORWALK VIRUS CAPSID | | Descriptor: | capsid protein | | Authors: | Prasad, B.V, Hardy, M.E, Dokland, T, Bella, J, Rossmann, M.G, Estes, M.K. | | Deposit date: | 2001-04-19 | | Release date: | 2001-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | X-ray crystallographic structure of the Norwalk virus capsid
Science, 286, 1999
|
|
6NIR
 
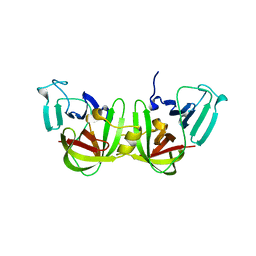 | | Crystal structure of a GII.4 norovirus HOV protease | | Descriptor: | HOV protease, HOV protease fragment | | Authors: | Prasad, B.V.V, Hu, L. | | Deposit date: | 2018-12-31 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | GII.4 Norovirus Protease Shows pH-Sensitive Proteolysis with a Unique Arg-His Pairing in the Catalytic Site.
J. Virol., 93, 2019
|
|
4IMZ
 
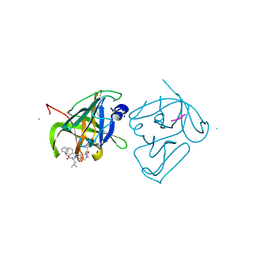 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | Genome polyprotein, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IN1
 
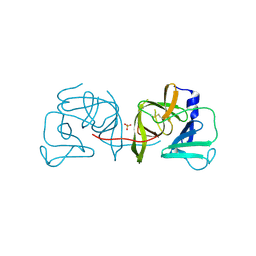 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | 3C-like protease, SULFATE ION | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IMQ
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | 3C-like protease, PEPTIDE INHIBITOR, syc8, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IN2
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | C-like protease | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4INH
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | DIMETHYL SULFOXIDE, Genome polyprotein, peptide inhibitor, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-04 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
8TL1
 
 | |
8TKA
 
 | |
8TL8
 
 | | Structure of Orthoreovirus RNA Chaperone SigmaNS R6A mutant in complex with bile acid | | Descriptor: | GLYCOCHOLIC ACID, Protein sigma-NS | | Authors: | Prasad, B.V.V, Zhao, B, Hu, L, Neetu, N. | | Deposit date: | 2023-07-26 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of orthoreovirus RNA chaperone sigma NS, a component of viral replication factories.
Nat Commun, 15, 2024
|
|
1UH8
 
 | |
1UH9
 
 | |
1UH7
 
 | |
1L9V
 
 | | Non Structural protein encoded by gene segment 8 of rotavirus (NSP2), an NTPase, ssRNA binding and nucleic acid helix-destabilizing protein | | Descriptor: | Rotavirus-NSP2 | | Authors: | Jayaram, H, Taraporewala, Z, Patton, J.T, Prasad, B.V. | | Deposit date: | 2002-03-26 | | Release date: | 2002-06-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rotavirus protein involved in genome replication and packaging exhibits a HIT-like fold.
Nature, 417, 2002
|
|
6AUK
 
 | |
6PCU
 
 | |
7RSW
 
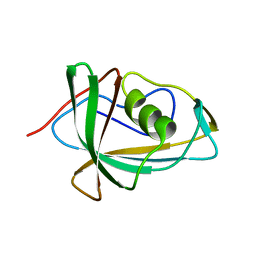 | | Crystal structure of group B human rotavirus VP8* | | Descriptor: | Outer capsid protein VP4, peptide | | Authors: | Hu, L, Salmen, W, Sankaran, B, Prasad, B.V. | | Deposit date: | 2021-08-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Novel fold of rotavirus glycan-binding domain predicted by AlphaFold2 and determined by X-ray crystallography.
Commun Biol, 5, 2022
|
|
2FYQ
 
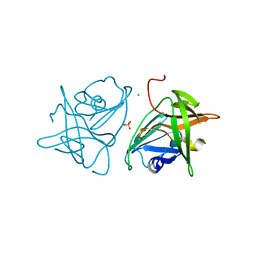 | |
2GE8
 
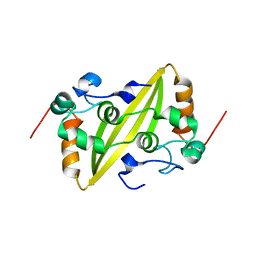 | | Structure of the C-terminal dimerization domain of infectious bronchitis virus nucleocapsid protein | | Descriptor: | Nucleocapsid protein | | Authors: | Jayaram, H, Fan, H, Bowman, B.R, Ooi, A, Jayaram, J, Collisson, E.W, Lescar, J, Prasad, B.V. | | Deposit date: | 2006-03-18 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation.
J.Virol., 80, 2006
|
|
2FYR
 
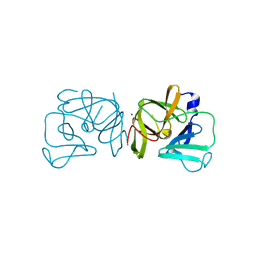 | |
2GEC
 
 | | Structure of the N-terminal domain of avian infectious bronchitis virus nucleocapsid protein (strain Gray) in a novel dimeric arrangement | | Descriptor: | Nucleocapsid protein | | Authors: | Jayaram, H, Fan, H, Bowman, B.R, Ooi, A, Jayaram, J, Collisson, E.W, Lescar, J, Prasad, B.V. | | Deposit date: | 2006-03-19 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation.
J.Virol., 80, 2006
|
|
2GE7
 
 | | Structure of the C-terminal dimerization domain of infectious bronchitis virus nucleocapsid protein | | Descriptor: | Nucleocapsid protein | | Authors: | Jayaram, H, Fan, H, Bowman, B.R, Ooi, A, Jayaram, J, Collisson, E.W, Lescar, J, Prasad, B.V. | | Deposit date: | 2006-03-18 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation.
J.Virol., 80, 2006
|
|
2GX9
 
 | |
5VX9
 
 | | VP8* of P[6] Human Rotavirus RV3 in complex with LNFP1 | | Descriptor: | Outer capsid protein VP4, SULFATE ION, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hu, L, Venkataram Prasad, B.V. | | Deposit date: | 2017-05-23 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Glycan recognition in globally dominant human rotaviruses.
Nat Commun, 9, 2018
|
|
5VX4
 
 | | VP8* of a G2P[4] Human Rotavirus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Outer capsid protein VP4 | | Authors: | Hu, L, Venkataram Prasad, B.V. | | Deposit date: | 2017-05-23 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Glycan recognition in globally dominant human rotaviruses.
Nat Commun, 9, 2018
|
|
