7ZTU
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform, D162L mutant | | Descriptor: | Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Silkworm carotenoprotein as an efficient carotenoid extractor, solubilizer and transporter.
Int.J.Biol.Macromol., 223, 2022
|
|
7ZVQ
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform, S206V mutant | | Descriptor: | Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the carotenoid binding and transport function of a START domain.
Structure, 30, 2022
|
|
7ZTQ
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform | | Descriptor: | Carotenoid-binding protein, GLYCEROL | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Silkworm carotenoprotein as an efficient carotenoid extractor, solubilizer and transporter.
Int.J.Biol.Macromol., 223, 2022
|
|
4FKC
 
 | | Recombinant prolidase from Thermococcus sibiricus | | Descriptor: | CADMIUM ION, Xaa-Pro aminopeptidase | | Authors: | Trofimov, A.A, Slutskaya, E.S, Polyakov, K.M, Dorovatovskii, P.V, Gumerov, V.M, Popov, V.O. | | Deposit date: | 2012-06-13 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Influence of intermolecular contacts on the structure of recombinant prolidase from Thermococcus sibiricus.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
8AIE
 
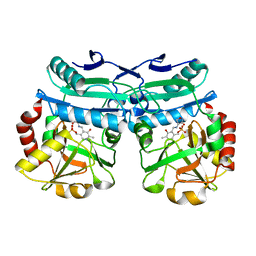 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense complexed with D-cycloserine | | Descriptor: | 3-azanyloxy-2-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]propanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class IV, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3D Structure of D-Аmino Acid Тransaminase from Aminobacterium colombiense in Complex with D-Cycloserine
Crystallography Reports, 68, 2023
|
|
8AHR
 
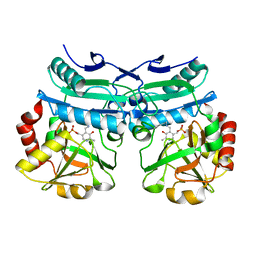 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense in holo form with PLP | | Descriptor: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-22 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense.
Molecules, 28, 2023
|
|
4H73
 
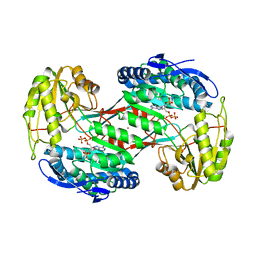 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. complexed with NADP+ | | Descriptor: | Aldehyde dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICKEL (II) ION | | Authors: | Petrova, T, Shabalin, I.G, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2012-09-20 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. complexed with NADP+
To be Published
|
|
4HA4
 
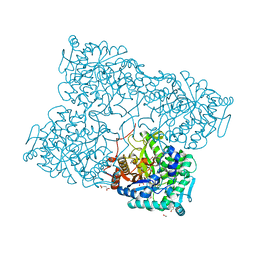 | | Structure of beta-glycosidase from Acidilobus saccharovorans in complex with glycerol | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Beta-galactosidase, GLYCEROL, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonov, A.V, Bezsudnova, E.Y, Dorovatovskii, P.V, Gumerov, V.M, Ravin, N.V, Skryabin, K.G, Popov, V.O. | | Deposit date: | 2012-09-25 | | Release date: | 2012-10-10 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structures of beta-glycosidase from Acidilobus saccharovorans in complexes with tris and glycerol.
Dokl.Biochem.Biophys., 449
|
|
7ZTR
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform, W232F mutant | | Descriptor: | Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Silkworm carotenoprotein as an efficient carotenoid extractor, solubilizer and transporter.
Int.J.Biol.Macromol., 223, 2022
|
|
8AYK
 
 | | Crystal structure of D-amino acid aminotrensferase from Aminobacterium colombiense complexed with D-glutamate | | Descriptor: | (~{Z})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pent-2-enedioic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class IV | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Minyaev, M.E, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense.
Molecules, 28, 2023
|
|
8AYJ
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiens complexed with 3-aminooxypropionic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
4HA3
 
 | | Structure of beta-glycosidase from Acidilobus saccharovorans in complex with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-galactosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonov, A.V, Bezsudnova, E.Y, Dorovatovskii, P.V, Gumerov, V.M, Ravin, N.V, Skryabin, K.G, Popov, V.O. | | Deposit date: | 2012-09-25 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structures of beta-glycosidase from Acidilobus saccharovorans in complexes with tris and glycerol.
Dokl.Biochem.Biophys., 449
|
|
4H05
 
 | | Crystal structure of aminoglycoside-3'-phosphotransferase of type VIII | | Descriptor: | Aminoglycoside-O-phosphotransferase VIII | | Authors: | Boyko, K.M, Gorbacheva, M.A, Danilenko, V.N, Alekseeva, M.G, Korzhenevskiy, D.A, Dorovatovskiy, P.V, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2012-09-07 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural characterization of the novel aminoglycoside phosphotransferase AphVIII from Streptomyces rimosus with enzymatic activity modulated by phosphorylation.
Biochem.Biophys.Res.Commun., 477, 2016
|
|
4EQ5
 
 | | DNA ligase from the archaeon Thermococcus sibiricus | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase | | Authors: | Petrova, T, Bezsudnova, E.Y, Dorokhov, B.D, Slutskaya, E.S, Polyakov, K.M, Dorovatovskiy, P.V, Ravin, N.V, Skryabin, K.G, Kovalchuk, M.V, Popov, V.O. | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Expression, purification, crystallization and preliminary crystallographic analysis of a thermostable DNA ligase from the archaeon Thermococcus sibiricus.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4HX0
 
 | | Crystal structure of a putative nucleotidyltransferase (TM1012) from Thermotoga maritima at 1.87 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,4-BUTANEDIOL, Putative nucleotidyltransferase TM1012, ... | | Authors: | Boyko, K.M, Gorbacheva, M.A, Korzhenevskiy, D.A, Lipkin, A.V, Popov, V.O, Kovalchuk, M.V, Shumilin, I.A, Minor, W, Shabalin, I.G, Golubev, A.M. | | Deposit date: | 2012-11-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of putative nucleotidyltransferase with two MPD molecules in the active site.
To be Published
|
|
4HGX
 
 | | Crystal structure of xylose isomerase domain containing protein (stm4435) from salmonella typhimurium lt2 with unknown ligand | | Descriptor: | ACETATE ION, Xylose isomerase domain containing protein, ZINC ION | | Authors: | Boyko, K.M, Gorbacheva, M.A, Korzhenevskiy, D.A, Dorovatovsky, P.V, Rakitina, T.V, Lipkin, A.V, Shumilin, I.A, Minor, W, Popov, V.O. | | Deposit date: | 2012-10-09 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of xylose isomerase domain containing protein (stm4435) from salmonella typhimurium lt2 with unknown ligand
To be Published
|
|
4NPO
 
 | | Crystal structure of protein with unknown function from Deinococcus radiodurans at P61 spacegroup | | Descriptor: | ACETATE ION, ACETYL GROUP, GLYCEROL, ... | | Authors: | Boyko, K.M, Gorbacheva, M.A, Rakitina, T.V, Korgenevsky, D.A, Shabalin, I.G, Shumilin, I.A, Dorovatovsky, P.V, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2013-11-22 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structure of protein with unknown function at P61 spacegroup
To be Published
|
|
4NS4
 
 | | Crystal structure of cold-active estarase from Psychrobacter cryohalolentis K5T | | Descriptor: | Alpha/beta hydrolase fold protein | | Authors: | Boyko, K.M, Petrovskaya, L.E, Gorbacheva, M.A, Korgenevsky, D.A, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2013-11-28 | | Release date: | 2015-01-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three-dimentional structure of an esterse from Psychrobacter cryohalolentis K5T provides clues to unusual thermostability of a cold-active enzyme
To be Published
|
|
4NMJ
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. complexed with NADP+ at 2 A resolution | | Descriptor: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Boyko, K.M, Bezsudnova, E.Y, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2013-11-15 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. complexed with NADP+ at 2 A resolution
To be Published
|
|
4NMK
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. crystallized in microgravity (complex with NADP+) | | Descriptor: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Boyko, K.M, Bezsudnova, E.Y, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2013-11-15 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. crystallized in microgravity (complex with NADP+)
To be Published
|
|
4NPA
 
 | | Scrystal structure of protein with unknown function from Vibrio cholerae at P22121 spacegroup | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Putative uncharacterized protein, ... | | Authors: | Boyko, K.M, Gorbacheva, M.A, Rakitina, T.V, Korgenevsky, D.A, Dorovatovsky, P.V, Lipkin, A.V, Minor, W, Shumilin, I.A, Popov, V.O. | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of protein with unknown function from Vibrio cholerae at P22121 spacegroup
To be Published
|
|
4R2W
 
 | | X-ray structure of uridine phosphorylase from Shewanella oneidensis MR-1 in complex with uridine at 1.6 A resolution | | Descriptor: | GLYCEROL, SULFATE ION, URIDINE, ... | | Authors: | Safonova, T.N, Mordkovich, N.N, Manuvera, V.A, Veiko, V.P, Popov, V.O, Polyakov, K.P. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-syn conformation of uridine and asymmetry of the hexameric molecule revealed in the high-resolution structures of Shewanella oneidensis MR-1 uridine phosphorylase in the free form and in complex with uridine.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4R2X
 
 | | Unique conformation of uridine and asymmetry of the hexameric molecule revealed in the high-resolution structures of Shewanella oneidensis uridine phosphorylase in the free form and in complex with uridine | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Safonova, T.N, Mordkovich, N.N, Manuvera, V.A, Veiko, V.P, Popov, V.O, Polyakov, K.M. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | High-syn conformation of uridine and asymmetry of the hexameric molecule revealed in the high-resolution structures of Shewanella oneidensis MR-1 uridine phosphorylase in the free form and in complex with uridine.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8YQ4
 
 | | Structure of mBaoJin2 | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Minyaev, M.E, Kuzmicheva, T.P, Vlaskina, A.V, Popov, V.O, Pyatkevich, K.D, Subach, F.V. | | Deposit date: | 2024-03-19 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of mBaoJin2
To Be Published
|
|
8C0N
 
 | | Crystal structure of the red form of the mTagFT fluorescent timer | | Descriptor: | Blue-to-red TagFT fluorescent timer | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Agapova, Y.K, Subach, O.M, Popov, V.O, Subach, F.V. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Blue-to-Red TagFT, mTagFT, mTsFT, and Green-to-FarRed mNeptusFT2 Proteins, Genetically Encoded True and Tandem Fluorescent Timers.
Int J Mol Sci, 24, 2023
|
|
