5L8Z
 
 | | Structure of thermostable DNA-binding HU protein from micoplasma Spiroplasma melliferum | | Descriptor: | DNA-binding protein, SODIUM ION | | Authors: | Boyko, K.M, Gorbacheva, M.A, Rakitina, T.V, Korzhenevskiy, D.A, Kamashev, D.E, Vanyushkina, A.A, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the high thermal stability of the histone-like HU protein from the mollicute Spiroplasma melliferum KC3.
Sci Rep, 6, 2016
|
|
8YTR
 
 | | The structure of Cu(II)-CopC from Thioalkalivibrio paradoxus | | Descriptor: | COPPER (II) ION, CopC domain-containing protein, DI(HYDROXYETHYL)ETHER | | Authors: | Kulikova, O.G, Solovieva, A.Y, Varfolomeeva, L.A, Dergousova, N.I, Nikolaeva, A.Y, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Cu(II)-CopC from Thioalkalivibrio paradoxus
To Be Published
|
|
8YTS
 
 | | The structure of the cytochrome c546/556 from Thioalkalivibrio paradoxus with unusual UV-Vis spectral features at atomic resolution | | Descriptor: | Cytochrome C, HEME C | | Authors: | Varfolomeeva, L.A, Solovieva, A.Y, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The structure of the cytochrome c546/556 from Thioalkalivibrio paradoxus with unusual UV-Vis spectral features at atomic resolution
To Be Published
|
|
8YU6
 
 | | The structure of thiocyanate dehydrogenase mutant with the H447Q substitution from Pelomicrobium methylotrophicum (pmTcDH H447Q), activated by crystal soaking with 1mM CuCl2 and 1 mM sodium ascorbate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Varfolomeeva, L.A, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structure of thiocyanate dehydrogenase mutant with the H447Q substitution from Pelomicrobium methylotrophicum (pmTcDH H447Q), activated by crystal soaking with 1mM CuCl2 and 1 mM sodium ascorbate
To Be Published
|
|
8YOU
 
 | | The pmTcDH complex structure with an inhibitor SeCN | | Descriptor: | COPPER (II) ION, GLYCEROL, SELENIUM ATOM, ... | | Authors: | Varfolomeeva, L.A, Polyakov, K.M, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-13 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The pmTcDH complex structure with an inhibitor SeCN
To Be Published
|
|
8YTQ
 
 | | The structure of apoCopC from Thioalkalivibrio paradoxus | | Descriptor: | ACETATE ION, COPPER (II) ION, CopC domain-containing protein, ... | | Authors: | Kulikova, O.G, Solovieva, A.Y, Varfolomeeva, L.A, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of apoCopC from Thioalkalivibrio paradoxus
To Be Published
|
|
8YU5
 
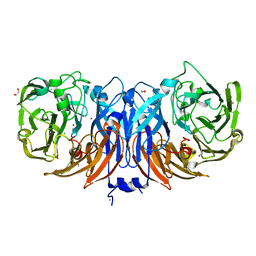 | | The structure of non-activated thiocyanate dehydrogenase mutant with the H447Q substitution from Pelomicrobium methylotrophicum (pmTcDH H447Q) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Varfolomeeva, L.A, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of non-activated thiocyanate dehydrogenase mutant with the H447Q substitution from Pelomicrobium methylotrophicum (pmTcDH H447Q)
To Be Published
|
|
8YRV
 
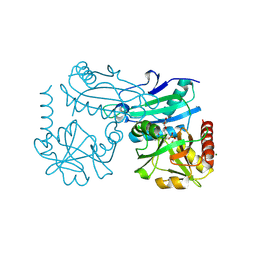 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis complexed with 3-aminooxypropionic acid | | Descriptor: | 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV, MAGNESIUM ION | | Authors: | Matyuta, I.O, Bakunova, A.K, Nikolaeva, A.Y, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-03-21 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis complexed with 3-aminooxypropionic acid
To Be Published
|
|
8YRT
 
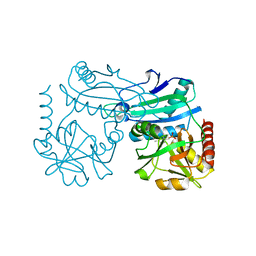 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis in the holo form obtained at pH 7.0 | | Descriptor: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-03-21 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Incorporation of pyridoxal-5'-phosphate into the apoenzyme: A structural study of D-amino acid transaminase from Haliscomenobacter hydrossis.
Biochim Biophys Acta Proteins Proteom, 1873, 2024
|
|
8YRU
 
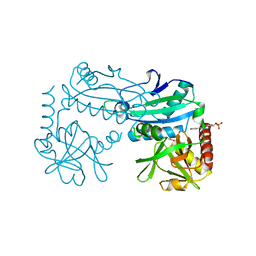 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis (apo form) after 15 sec of soaking with phenylhydrazine | | Descriptor: | ACETATE ION, Aminotransferase class IV, [6-methyl-5-oxidanyl-4-[(2-phenylhydrazinyl)methyl]pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-03-21 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Incorporation of pyridoxal-5'-phosphate into the apoenzyme: A structural study of D-amino acid transaminase from Haliscomenobacter hydrossis.
Biochim Biophys Acta Proteins Proteom, 1873, 2024
|
|
8ZBO
 
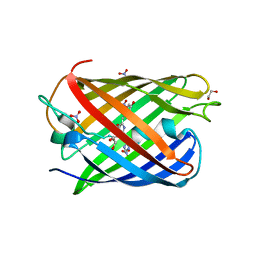 | | Crystal structure of the biphotochromic fluorescent protein moxSAASoti (F97M variant) in its green on-state | | Descriptor: | 1,2-ETHANEDIOL, F97M variant of the biphotochromic fluorescent protein moxSAASoti, NITRATE ION, ... | | Authors: | Boyko, K.M, Matyuta, I.O, Marynich, N.K, Minyaev, M.E, Khadiyatova, A.A, Popov, V.O, Savitsky, A.P. | | Deposit date: | 2024-04-26 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single-point substitution F97M leads to in cellulo crystallization of the biphotochromic protein moxSAASoti.
Biochem.Biophys.Res.Commun., 732, 2024
|
|
6ET6
 
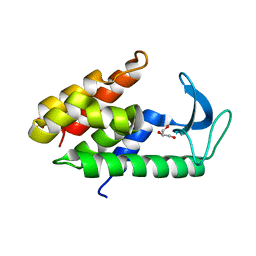 | | Crystal structure of muramidase from Acinetobacter baumannii AB 5075UW prophage | | Descriptor: | GLYCEROL, Lysozyme, SULFATE ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Sykilinda, N.N, Shneider, M.M, Miroshnikov, K.A, Popov, V.O. | | Deposit date: | 2017-10-25 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of anAcinetobacterBroad-Range Prophage Endolysin Reveals a C-Terminal alpha-Helix with the Proposed Role in Activity against Live Bacterial Cells.
Viruses, 10, 2018
|
|
6FUC
 
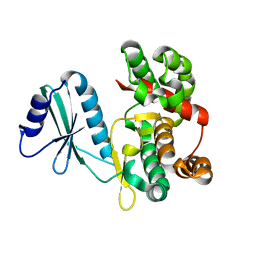 | | Structure of aminoglycoside phosphotransferase APH(3'')-Id from Streptomyces rimosus ATCC10970 | | Descriptor: | Aminoglycoside phosphotransferase | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Alekseeva, M.G, Mavletova, D.A, Zakharevich, N.V, Rudakova, N.N, Danilenko, V.N, Popov, V.O. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Identification, functional and structural characterization of novel aminoglycoside phosphotransferase APH(3′′)-Id from Streptomyces rimosus subsp. rimosus ATCC 10970.
Arch.Biochem.Biophys., 671, 2019
|
|
6FUX
 
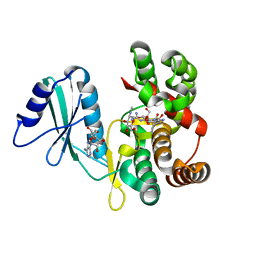 | | Structure of aminoglycoside phosphotransferase APH(3'')-Id from Streptomyces rimosus ATCC10970 in complex with ADP and streptomycin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aminoglycoside phosphotransferase, GLYCEROL, ... | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Alekseeva, M.G, Mavletova, D.A, Zakharevich, N.V, Rudakova, N.N, Danilenko, V.N, Popov, V.O. | | Deposit date: | 2018-02-28 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification, functional and structural characterization of novel aminoglycoside phosphotransferase APH(3′′)-Id from Streptomyces rimosus subsp. rimosus ATCC 10970.
Arch.Biochem.Biophys., 671, 2019
|
|
6GKR
 
 | | Crystal structure of branched-chain amino acid aminotransferase from Thermobaculum terrenum in PLP-form (holo-form) | | Descriptor: | ACETATE ION, Branched-chain-amino-acid aminotransferase, CHLORIDE ION, ... | | Authors: | Boyko, K.M, Bezsudnova, E.Y, Nikolaeva, A.Y, Zeifman, Y.S, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2018-05-21 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Biochemical and structural insights into PLP fold type IV transaminase from Thermobaculum terrenum.
Biochimie, 158, 2018
|
|
6G5M
 
 | |
6H65
 
 | | Crystal structure of the branched-chain-amino-acid aminotransferase from Haliangium ochraceum | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Timofeev, V.I, Bezsudnova, E.Y, Nikolaeva, A.Y, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2018-07-26 | | Release date: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the branched-chain-amino-acid aminotransferase from Haliangium ochraceum
To Be Published
|
|
3RKH
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with nitrite (full occupancy) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonova, T.V, Tikhonov, A.V, Dorovatovskii, P.V, Popov, V.O. | | Deposit date: | 2011-04-18 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6G50
 
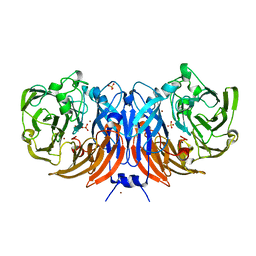 | | The structure of thiocyanate dehydrogenase from Thioalkalivibrio paradoxus as isolated. | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, SULFATE ION, ... | | Authors: | Polyakov, K.M, Tsallagov, S.I, Tikhkonova, T.V, Popov, V.O. | | Deposit date: | 2018-03-28 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Trinuclear copper biocatalytic center forms an active site of thiocyanate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3RR5
 
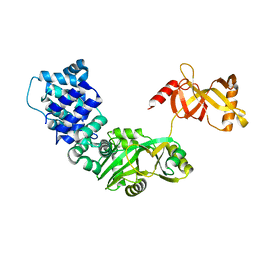 | | DNA ligase from the archaeon Thermococcus sp. 1519 | | Descriptor: | DNA ligase, MAGNESIUM ION | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Popov, V.O, Polyakov, K.M, Ravin, N.V, Shabalin, I.G, Skryabin, K.G, Stekhanova, T.N, Kovalchuk, M.V. | | Deposit date: | 2011-04-29 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.018 Å) | | Cite: | ATP-dependent DNA ligase from Thermococcus sp. 1519 displays a new arrangement of the OB-fold domain.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3OWM
 
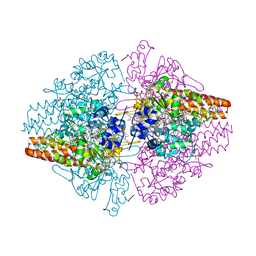 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with hydroxylamine | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Tikhonova, T.V, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2010-09-20 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding of sulfite by the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase
To be Published
|
|
3S7W
 
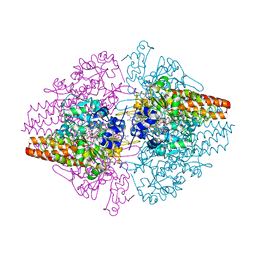 | | Structure of the TvNiRb form of Thioalkalivibrio nitratireducens cytochrome c nitrite reductase with an oxidized Gln360 in a complex with hydroxylamine | | Descriptor: | AZIDE ION, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2011-05-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of the TvNiRb form of Thioalkalivibrio nitratireducens cytochrome c nitrite reductase with an oxidized Gln360 in a complex with hydroxylamine
To be Published
|
|
3SCE
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase with a covalent bond between the CE1 atom of Tyr303 and the CG atom of Gln360 (TvNiRb) | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2011-06-07 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7P8O
 
 | | Crystal structure of D-aminoacid transaminase from Haliscomenobacter hydrossis in its intermediate form | | Descriptor: | Aminotransferase class IV, MAGNESIUM ION, SULFATE ION | | Authors: | Matyuta, I.O, Boyko, K.M, Bakunova, A.K, Nikolaeva, A.Y, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2021-07-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Incorporation of pyridoxal-5'-phosphate into the apoenzyme: A structural study of D-amino acid transaminase from Haliscomenobacter hydrossis.
Biochim Biophys Acta Proteins Proteom, 1873, 2024
|
|
5CE8
 
 | | Crystal structure of branched-chain aminotransferase from thermophilic archaea Thermoproteus uzoniensis | | Descriptor: | Branched-chain amino acid aminotransferase, DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Stekhanova, T.N, Mardanov, A.V, Rakitin, A.L, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-07-06 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | First structure of archaeal branched-chain amino acid aminotransferase from Thermoproteus uzoniensis specific for L-amino acids and R-amines.
Extremophiles, 20, 2016
|
|
