1X93
 
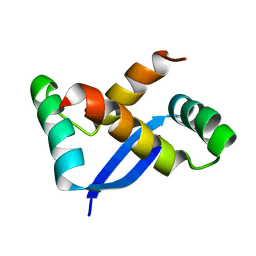 | | NMR Structure of Helicobacter pylori HP0222 | | Descriptor: | hypothetical protein HP0222 | | Authors: | Popescu, A, Karpay, A, Israel, D, Peek Jr, R.M, Krezel, A.M. | | Deposit date: | 2004-08-19 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Helicobacter pylori protein HP0222 belongs to Arc/MetJ family of transcriptional regulators.
Proteins, 59, 2005
|
|
2Y0C
 
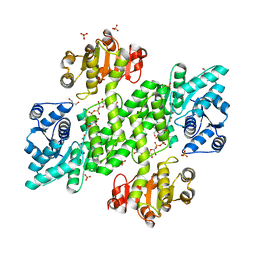 | | BceC mutation Y10S | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Rocha, J, Popescu, A.O, Borges, P, Mil-Homens, D, Sa-Correia, I, Fialho, A.M, Frazao, C. | | Deposit date: | 2010-12-02 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of Burkholderia Cepacia Udp-Glucose Dehydrogenase (Ugd) Bcec and Role of Tyr10 in Final Hydrolysis of Ugd Thioester Intermediate.
J.Bacteriol., 193, 2011
|
|
2Y0E
 
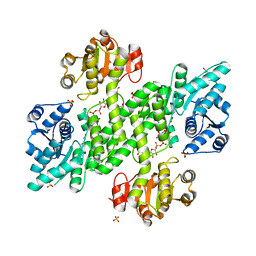 | | BceC and the final step of UGDs reaction | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Rocha, J, Popescu, A.O, Borges, P, Mil-Homens, D, Sa-Correia, I, Fialho, A.M, Frazao, C. | | Deposit date: | 2010-12-02 | | Release date: | 2011-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of Burkholderia Cepacia Udp-Glucose Dehydrogenase (Ugd) Bcec and Role of Tyr10 in Final Hydrolysis of Ugd Thioester Intermediate.
J.Bacteriol., 193, 2011
|
|
2Y0D
 
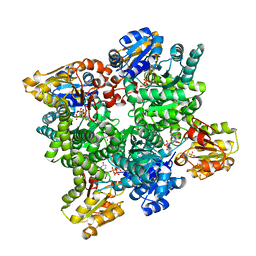 | | BceC mutation Y10K | | Descriptor: | SULFATE ION, UDP-GLUCOSE DEHYDROGENASE, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Rocha, J, Popescu, A.O, Borges, P, Mil-Homens, D, Sa-Correia, I, Fialho, A.M, Frazao, C. | | Deposit date: | 2010-12-02 | | Release date: | 2011-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Burkholderia Cepacia Udp-Glucose Dehydrogenase (Ugd) Bcec and Role of Tyr10 in Final Hydrolysis of Ugd Thioester Intermediate.
J.Bacteriol., 193, 2011
|
|
1ROT
 
 | | STRUCTURE OF FKBP59-I, THE N-TERMINAL DOMAIN OF A 59 KDA FK506-BINDING PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FKBP59-I | | Authors: | Craescu, C.T, Rouviere, N, Popescu, A, Cerpolini, E, Lebeau, M.-C, Baulieu, E.-E, Mispelter, J. | | Deposit date: | 1996-06-14 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the immunophilin-like domain of FKBP59 in solution.
Biochemistry, 35, 1996
|
|
1ROU
 
 | | STRUCTURE OF FKBP59-I, THE N-TERMINAL DOMAIN OF A 59 KDA FK506-BINDING PROTEIN, NMR, 22 STRUCTURES | | Descriptor: | FKBP59-I | | Authors: | Craescu, C.T, Rouviere, N, Popescu, A, Cerpolini, E, Lebeau, M.-C, Baulieu, E.-E, Mispelter, J. | | Deposit date: | 1996-06-14 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the immunophilin-like domain of FKBP59 in solution.
Biochemistry, 35, 1996
|
|
