2VQX
 
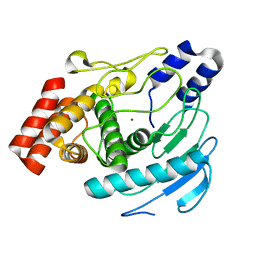 | | Precursor of Protealysin, Metalloproteinase from Serratia proteamaculans. | | Descriptor: | METALLOPROTEINASE, ZINC ION | | Authors: | Melik-Adamyan, W.R, Kuranova, I.P, Polyakov, K.M, Demidyuk, I.V, Gromova, T.Y, Kostrov, S.V. | | Deposit date: | 2008-03-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Crystal Structure of the Protealysin Precursor: Insights Into Propeptide Function.
J.Biol.Chem., 285, 2010
|
|
1OBR
 
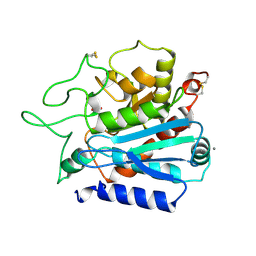 | | CARBOXYPEPTIDASE T | | Descriptor: | CALCIUM ION, CARBOXYPEPTIDASE T, SULFATE ION, ... | | Authors: | Teplyakov, A, Polyakov, K, Obmolova, G, Osterman, A. | | Deposit date: | 1996-06-22 | | Release date: | 1997-01-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of carboxypeptidase T from Thermoactinomyces vulgaris.
Eur.J.Biochem., 208, 1992
|
|
2GUG
 
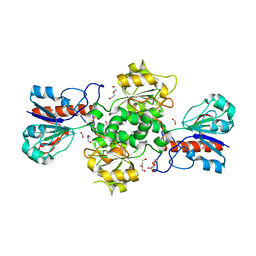 | | NAD-dependent formate dehydrogenase from Pseudomonas sp.101 in complex with formate | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, Formate dehydrogenase, ... | | Authors: | Filippova, E.V, Polyakov, K.M, Tikhonova, T.V, Boiko, K.M, Tishkov, V.I, Popov, V.O. | | Deposit date: | 2006-04-30 | | Release date: | 2006-05-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the complex of NAD-dependent formate dehydrogenase from
metylotrophic bacterium Pseudomonas sp.101 with formate.
KRISTALLOGRAFIYA, 51, 2006
|
|
7Q1J
 
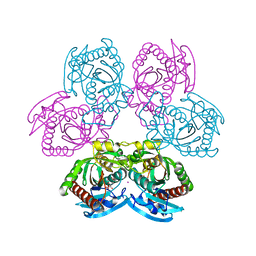 | |
7Q32
 
 | | Mutant D24G of uridine phosphorylase from E. coli | | Descriptor: | CITRATE ANION, POTASSIUM ION, Uridine phosphorylase | | Authors: | Safonova, T, Polyakov, K, Antipov, A, Okorokova, N, Mordkovich, N, Veiko, V. | | Deposit date: | 2021-10-26 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mutant D24G of uridine phosphorylase from E. coli
To Be Published
|
|
2GO1
 
 | | NAD-dependent formate dehydrogenase from Pseudomonas sp.101 | | Descriptor: | NAD-dependent formate dehydrogenase, SULFATE ION | | Authors: | Filippova, E.V, Polyakov, K.M, Tikhonova, T.V, Stekhanova, T.N, Boiko, K.M, Popov, V.O. | | Deposit date: | 2006-04-12 | | Release date: | 2006-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a new crystal modification of the bacterial NAD-dependent formate dehydrogenase with a resolution of 2.1 A
Crystallography reports, 50, 2005
|
|
1M38
 
 | | Structure of Inorganic Pyrophosphatase | | Descriptor: | COBALT (II) ION, INORGANIC PYROPHOSPHATASE, PHOSPHATE ION | | Authors: | Kuranova, I.P, Polyakov, K.M, Levdikov, V.M, Smirnova, E.A, Hohne, W.E, Lamzin, V.S, Meijers, R. | | Deposit date: | 2002-06-27 | | Release date: | 2003-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of Saccharomyces cerevisiae inorganic pyrophosphatase complexed with cobalt and phosphate ions
CRYSTALLOGRAPHY REPORTS, 48, 2003
|
|
3UU9
 
 | | Structure of the free TvNiRb form of Thioalkalivibrio nitratireducens cytochrome c nitrite reductase | | Descriptor: | CALCIUM ION, Eight-heme nitrite reductase, HEME C, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonova, T.V, Tikhonov, A.V, Dorovatovskii, P.V, Popov, V.O. | | Deposit date: | 2011-11-28 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2GSD
 
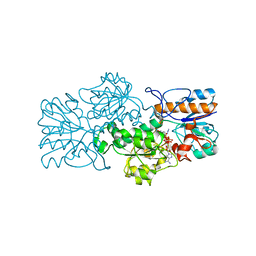 | | NAD-dependent formate dehydrogenase from bacterium Moraxella sp.C2 in complex with NAD and azide | | Descriptor: | AZIDE ION, NAD-dependent formate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Filippova, E.V, Polyakov, K.M, Tikhonova, T.V, Sadykhov, I.G, Shabalin, I.G, Tishkov, V.I, Popov, V.O. | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the apo and holo forms of formate dehydrogenase from the bacterium Moraxella sp. C-1: towards understanding the mechanism of the closure of the interdomain cleft.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4R2W
 
 | | X-ray structure of uridine phosphorylase from Shewanella oneidensis MR-1 in complex with uridine at 1.6 A resolution | | Descriptor: | GLYCEROL, SULFATE ION, URIDINE, ... | | Authors: | Safonova, T.N, Mordkovich, N.N, Manuvera, V.A, Veiko, V.P, Popov, V.O, Polyakov, K.P. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-syn conformation of uridine and asymmetry of the hexameric molecule revealed in the high-resolution structures of Shewanella oneidensis MR-1 uridine phosphorylase in the free form and in complex with uridine.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5EK6
 
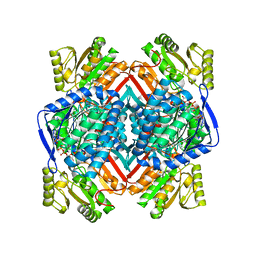 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. 1860 complexed with NADP and isobutyraldehyde | | Descriptor: | 2-methylpropanal, Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T.E, Bezsudnova, E.Y, Boyko, K.M, Polyakov, K.M, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2015-11-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
4L3Z
 
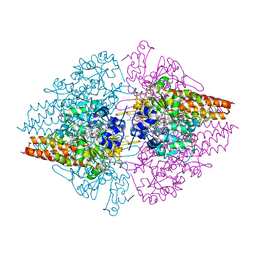 | | Nitrite complex of TvNiR, second middle dose data set | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Lazarenko, V.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2013-06-07 | | Release date: | 2014-06-11 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Investigation of the X-ray-induced nitrite reduction catalysed by cytochrome c nitrite reductase from the bacterium Thioalkalivibrio nitratireducens
To be Published
|
|
4L38
 
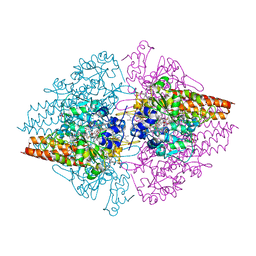 | | Nitrite complex of TvNiR, low dose data set | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Lazarenko, V.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2013-06-05 | | Release date: | 2014-07-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Investigation of the X-ray-induced nitrite reduction catalysed by cytochrome c nitrite reductase from the bacterium Thioalkalivibrio nitratireducens
To be Published
|
|
4L3Y
 
 | | Nitrite complex of TvNiR, high dose data set (NO complex) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Lazarenko, V.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2013-06-07 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Investigation of the X-ray-induced nitrite reduction catalysed by cytochrome c nitrite reductase from the bacterium Thioalkalivibrio nitratireducens
To be Published
|
|
4L3X
 
 | | Nitrite complex of TvNiR, first middle dose data set | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Lazarenko, V.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2013-06-07 | | Release date: | 2014-06-11 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Investigation of the X-ray-induced nitrite reduction catalysed by cytochrome c nitrite reductase from the bacterium Thioalkalivibrio nitratireducens
To be Published
|
|
4HA4
 
 | | Structure of beta-glycosidase from Acidilobus saccharovorans in complex with glycerol | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Beta-galactosidase, GLYCEROL, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonov, A.V, Bezsudnova, E.Y, Dorovatovskii, P.V, Gumerov, V.M, Ravin, N.V, Skryabin, K.G, Popov, V.O. | | Deposit date: | 2012-09-25 | | Release date: | 2012-10-10 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structures of beta-glycosidase from Acidilobus saccharovorans in complexes with tris and glycerol.
Dokl.Biochem.Biophys., 449
|
|
3N7U
 
 | | NAD-dependent formate dehydrogenase from higher-plant Arabidopsis thaliana in complex with NAD and azide | | Descriptor: | AZIDE ION, Formate dehydrogenase, GLYCEROL, ... | | Authors: | Shabalin, I.G, Polyakov, K.M, Serov, A.E, Skirgello, O.E, Sadykhov, E.G, Dorovatovskiy, P.V, Tishkov, V.I, Popov, V.O. | | Deposit date: | 2010-05-27 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the apo and holo forms of NAD-dependent formate dehydrogenase from the higher-plant Arabidopsis Thaliana
To be Published
|
|
3LGQ
 
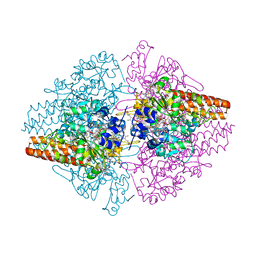 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in complex with sulfite (modified Tyr-303) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Filimonenkov, A.A, Tikhonova, T.V, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2010-01-21 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3LG1
 
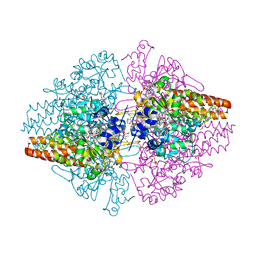 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase reduced by sodium borohydride (in complex with sulfite) | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Filimonenkov, A.A, Dorovatovsky, P.V, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2010-01-19 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3MMO
 
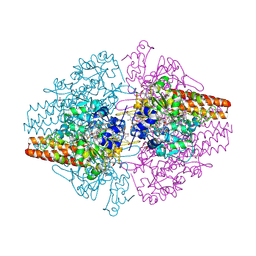 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in complex with cyanide | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Tikhonova, T.V, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2010-04-20 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of complexes of octahaem cytochrome c nitrite reductase from Thioalkalivibrio nitratireducens with sulfite and cyanide.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4HA3
 
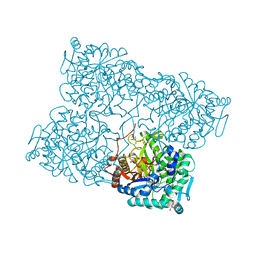 | | Structure of beta-glycosidase from Acidilobus saccharovorans in complex with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-galactosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonov, A.V, Bezsudnova, E.Y, Dorovatovskii, P.V, Gumerov, V.M, Ravin, N.V, Skryabin, K.G, Popov, V.O. | | Deposit date: | 2012-09-25 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structures of beta-glycosidase from Acidilobus saccharovorans in complexes with tris and glycerol.
Dokl.Biochem.Biophys., 449
|
|
3GM6
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in complex with phosphate | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Filimonenkov, A.A, Dorovatovsky, P.V, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2009-03-13 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of octaheme cytochrome c nitrite reductase from Thioalkalivibrio nitratireducens in a complex with phosphate
Crystallography Reports, 55, 2010
|
|
3FO3
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase reduced by sodium dithionite (sulfite complex) | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-12-27 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of complexes of octahaem cytochrome c nitrite reductase from Thioalkalivibrio nitratireducens with sulfite and cyanide
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5F75
 
 | | Thiocyanate dehydrogenase from Thioalkalivibrio paradoxus | | Descriptor: | COPPER (II) ION, Thiocyanate dehydrogenase | | Authors: | Tsallagov, S.I, Polyakov, K.M, Tikhonova, T.V, Trofimov, A.A, Shabalin, I.G, Popov, A.N, Popov, V.O. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thiocyanate dehydrogenase from Thioalkalivibrio paradoxus
To Be Published
|
|
3OWM
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with hydroxylamine | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Tikhonova, T.V, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2010-09-20 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding of sulfite by the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase
To be Published
|
|
