2OGR
 
 | |
4ZFS
 
 | |
4EDS
 
 | |
4EDO
 
 | |
3PJ5
 
 | |
3PJB
 
 | |
4HE4
 
 | |
4N3D
 
 | | Crystal structure of the dimeric variant EGFP-K162Q in P61 space group | | Descriptor: | GLYCEROL, Green fluorescent protein, PHOSPHATE ION, ... | | Authors: | Pletneva, N.V, Pletnev, V.Z, Pletnev, S.V. | | Deposit date: | 2013-10-07 | | Release date: | 2014-08-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Three dimensional structure of the dimeric gene-engineered variant of green fluorescent protein egfp-K162Q in P61 crystal space group
Rus.J.Bioorg.Chem., 40, 2014
|
|
3PIB
 
 | |
3PJ7
 
 | |
3LVD
 
 | |
3LVC
 
 | |
3LVA
 
 | |
3GL4
 
 | |
3GB3
 
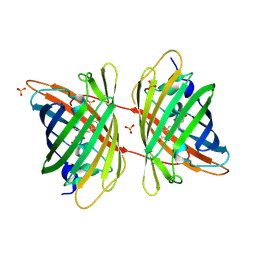 | |
2BQP
 
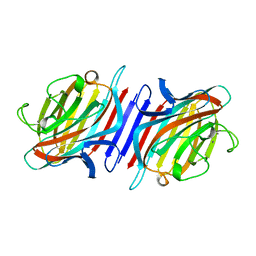 | | THE STRUCTURE OF THE PEA LECTIN-D-GLUCOPYRANOSE COMPLEX | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEA LECTIN), ... | | Authors: | Ruzeinikov, S.N, Mikhailova Yu, I, Tsygannik, I.N, Pangborn, W, Duax, W, Pletnev, V.Z. | | Deposit date: | 1998-12-08 | | Release date: | 1998-12-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of Pea Lectin-D-Glucopyranose Complex at a 1.9 A Resolution
RUSS.J.BIOORGANIC CHEM., 23, 1997
|
|
1F90
 
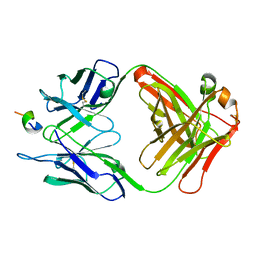 | | FAB FRAGMENT OF MONOCLONAL ANTIBODY (LNKB-2) AGAINST HUMAN INTERLEUKIN-2 IN COMPLEX WITH ANTIGENIC PEPTIDE | | Descriptor: | ANTIGENIC NONAPEPTIDE, FAB FRAGMENT OF MONOCLONAL ANTIBODY | | Authors: | Afonin, P.V, Fokin, A.V, Tsigannik, I.N, Mikhailova, I.Y, Onoprienko, L.V, Mikhaleva, I.I, Ivanov, V.T, Mareeva, T.Y, Nesmeyanov, V.A, Li, N, Duax, W.L, Pletnev, V.Z. | | Deposit date: | 2000-07-06 | | Release date: | 2001-07-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an anti-interleukin-2 monoclonal antibody Fab complexed with an antigenic nonapeptide.
Protein Sci., 10, 2001
|
|
1BQP
 
 | | THE STRUCTURE OF THE PEA LECTIN-D-MANNOPYRANOSE COMPLEX | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (LECTIN), ... | | Authors: | Ruzeinikov, S.N, Mikhailova, I.Y, Tsygannik, I.N, Pangborn, W, Duax, W, Pletnev, V.Z. | | Deposit date: | 1998-08-17 | | Release date: | 1998-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the Pea Lectin-D-Mannopyranose Complex at a 2.1 A Resolution
RUSS.J.BIOORGANIC CHEM., 24, 1998
|
|
