8CO1
 
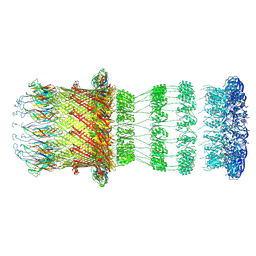 | | Type II Secretion System | | Descriptor: | IPT/TIG domain-containing protein, Lipoprotein, Probable type IV piliation system protein DR_0774 | | Authors: | Farci, D, Piano, D. | | Deposit date: | 2023-02-26 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural characterization and functional insights into the type II secretion system of the poly-extremophile Deinococcus radiodurans.
J.Biol.Chem., 300, 2024
|
|
7ZGX
 
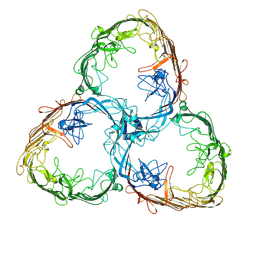 | | S-layer Deinoxanthin Binding Complex, C1 symmetry | | Descriptor: | S-layer protein SlpA | | Authors: | Farci, D, Piano, D. | | Deposit date: | 2022-04-04 | | Release date: | 2022-07-13 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | The cryo-EM structure of the S-layer deinoxanthin-binding complex of Deinococcus radiodurans informs properties of its environmental interactions.
J.Biol.Chem., 298, 2022
|
|
7ZGY
 
 | | S-layer Deinoxanthin Binding Complex, C3 symmetry | | Descriptor: | (3~{S},5~{R},6~{R})-5-[(3~{S},7~{R},12~{S},16~{S},20~{S})-3,7,12,16,20,24-hexamethyl-24-oxidanyl-pentacosyl]-4,4,6-trimethyl-cyclohexane-1,3-diol, COPPER (II) ION, FE (III) ION, ... | | Authors: | Farci, D, Piano, D. | | Deposit date: | 2022-04-04 | | Release date: | 2022-07-13 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | The cryo-EM structure of the S-layer deinoxanthin-binding complex of Deinococcus radiodurans informs properties of its environmental interactions.
J.Biol.Chem., 298, 2022
|
|
8ACQ
 
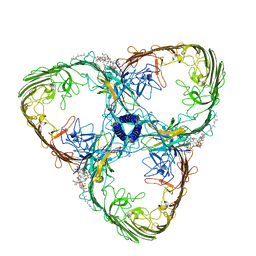 | | S-layer Deinoxanthin-Binding Complex (SDBC), subunit DR_2577 assembled with its SOD DR_0644 | | Descriptor: | (3~{S},5~{R},6~{R})-5-[(3~{S},7~{R},12~{S},16~{S},20~{S})-3,7,12,16,20,24-hexamethyl-24-oxidanyl-pentacosyl]-4,4,6-trimethyl-cyclohexane-1,3-diol, COPPER (II) ION, DR_0644, ... | | Authors: | Farci, D, Piano, D. | | Deposit date: | 2022-07-06 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | The SDBC is active in quenching oxidative conditions and bridges the cell envelope layers in Deinococcus radiodurans.
J.Biol.Chem., 299, 2023
|
|
8ACA
 
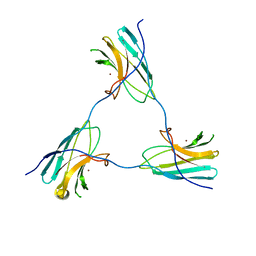 | | SDBC DR_0644 subunit, only-Cu Superoxide Dismutase | | Descriptor: | COPPER (II) ION, DR_0644, only-Cu Superoxide Dismutase | | Authors: | Farci, D, Piano, D. | | Deposit date: | 2022-07-05 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | The SDBC is active in quenching oxidative conditions and bridges the cell envelope layers in Deinococcus radiodurans.
J.Biol.Chem., 299, 2023
|
|
8AGD
 
 | | Full SDBC and SOD assembly | | Descriptor: | (3~{S},5~{R},6~{R})-5-[(3~{S},7~{R},12~{S},16~{S},20~{S})-3,7,12,16,20,24-hexamethyl-24-oxidanyl-pentacosyl]-4,4,6-trimethyl-cyclohexane-1,3-diol, COPPER (II) ION, FE (III) ION, ... | | Authors: | Farci, D, Graca, A.T, Piano, D. | | Deposit date: | 2022-07-19 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The SDBC is active in quenching oxidative conditions and bridges the cell envelope layers in Deinococcus radiodurans.
J.Biol.Chem., 299, 2023
|
|
7NWR
 
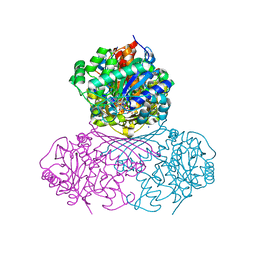 | | Structure of BT1526, a myo-inositol-1-phosphate synthase | | Descriptor: | Inositol-3-phosphate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Basle, A, Tang, G, Marles-Wright, J, Campopiano, D. | | Deposit date: | 2021-03-17 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of inositol lipid metabolism in gut-associated Bacteroidetes.
Nat Microbiol, 7, 2022
|
|
1DTY
 
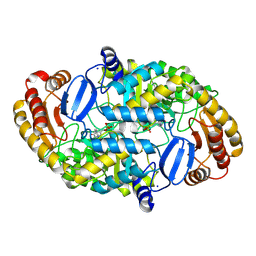 | | CRYSTAL STRUCTURE OF ADENOSYLMETHIONINE-8-AMINO-7-OXONANOATE AMINOTRANSFERASE WITH PYRIDOXAL PHOSPHATE COFACTOR. | | Descriptor: | ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION | | Authors: | Alexeev, D, Sawyer, L, Baxter, R.L, Alexeeva, M.V, Campopiano, D. | | Deposit date: | 2000-01-13 | | Release date: | 2000-02-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of adenosylmethionine-8-amino-7-oxonanoate aminotransferase with pyridoxal phosphate cofactor
to be published
|
|
5FJP
 
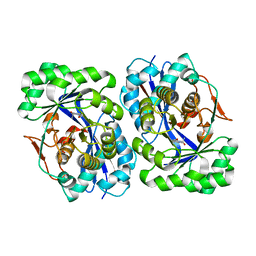 | |
5FJR
 
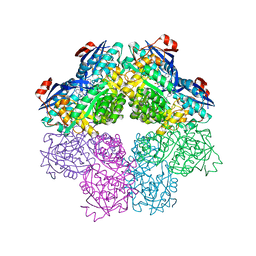 | |
5FJU
 
 | |
5FJT
 
 | |
5FJO
 
 | |
7POA
 
 | | An Irreversible, Promiscuous and Highly Thermostable Claisen-Condensation Biocatalyst Drives the Synthesis of Substituted Pyrroles | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-amino-7-oxononanoate synthase/2-amino-3-ketobutyrate coenzyme A ligase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Basle, A, Ashley, B, Campopiano, D, Marles-Wright, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Versatile Chemo-Biocatalytic Cascade Driven by a Thermophilic and Irreversible C-C Bond-Forming alpha-Oxoamine Synthase.
Acs Sustain Chem Eng, 11, 2023
|
|
7POB
 
 | | An Irreversible, Promiscuous and Highly Thermostable Claisen-Condensation Biocatalyst Drives the Synthesis of Substituted Pyrroles | | Descriptor: | 8-amino-7-oxononanoate synthase/2-amino-3-ketobutyrate coenzyme A ligase, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION | | Authors: | Basle, A, Ashley, B, Campopiano, D, Marles-Wright, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Versatile Chemo-Biocatalytic Cascade Driven by a Thermophilic and Irreversible C-C Bond-Forming alpha-Oxoamine Synthase.
Acs Sustain Chem Eng, 11, 2023
|
|
7POC
 
 | | An Irreversible, Promiscuous and Highly Thermostable Claisen-Condensation Biocatalyst Drives the Synthesis of Substituted Pyrroles | | Descriptor: | 8-amino-7-oxononanoate synthase/2-amino-3-ketobutyrate coenzyme A ligase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Basle, A, Ashley, B, Campopiano, D, Marles-Wright, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Versatile Chemo-Biocatalytic Cascade Driven by a Thermophilic and Irreversible C-C Bond-Forming alpha-Oxoamine Synthase.
Acs Sustain Chem Eng, 11, 2023
|
|
3FVQ
 
 | | Crystal structure of the nucleotide binding domain FbpC complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Fe(3+) ions import ATP-binding protein fbpC | | Authors: | Newstead, S, Bilton, P, Carpenter, E.P, Campopiano, D, Iwata, S. | | Deposit date: | 2009-01-16 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into how nucleotide-binding domains power ABC transport.
Structure, 17, 2009
|
|
