4W79
 
 | | Crystal Structure of Human Protein N-terminal Glutamine Amidohydrolase | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, Protein N-terminal glutamine amidohydrolase, ... | | Authors: | Bitto, E, Bingman, C.A, McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human protein N-terminal glutamine amidohydrolase, an initial component of the N-end rule pathway.
Plos One, 9, 2014
|
|
3BUS
 
 | | Crystal Structure of RebM | | Descriptor: | Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McCoy, J.G, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2008-01-03 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and mechanism of the rebeccamycin sugar 4'-O-methyltransferase RebM.
J.Biol.Chem., 283, 2008
|
|
3BZB
 
 | | Crystal structure of uncharacterized protein CMQ451C from the primitive red alga Cyanidioschyzon merolae | | Descriptor: | Uncharacterized protein | | Authors: | McCoy, J.G, Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-01-17 | | Release date: | 2008-01-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of uncharacterized protein CMQ451C from the primitive red alga Cyanidioschyzon merolae.
To be Published
|
|
3DCY
 
 | |
3DHI
 
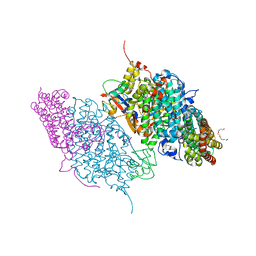 | | Crystal Structure of Reduced Toluene 4-Monoxygenase Hydroxylase Complexed with Effector Protein | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, FE (III) ION, ... | | Authors: | Bailey, L.J, Mccoy, J.G, Phillips Jr, G.N, Fox, B.G. | | Deposit date: | 2008-06-17 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural consequences of effector protein complex formation in a diiron hydroxylase.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3DHH
 
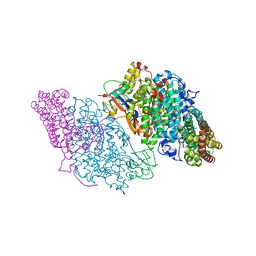 | | Crystal Structure of Resting State Toluene 4-Monoxygenase Hydroxylase Complexed with Effector Protein | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-BROMOPHENOL, CHLORIDE ION, ... | | Authors: | Bailey, L.J, Mccoy, J.G, Phillips Jr, G.N, Fox, B.G. | | Deposit date: | 2008-06-17 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural consequences of effector protein complex formation in a diiron hydroxylase.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
5MG0
 
 | | Structure of PAS-GAF fragment of Deinococcus phytochrome by serial femtosecond crystallography | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, ... | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Miller, M.D, Young, I.D, Brewster, A.S, Clinger, J, Aller, P, Braeuer, P, Hutchison, C, Alonso-Mori, R, Kern, J, Yachandra, V.K, Yano, J, Sauter, N.K, Phillips Jr, G.N, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
3H7C
 
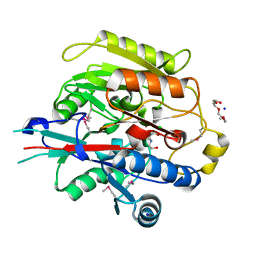 | | Crystal Structure of Arabidopsis thaliana Agmatine Deiminase from Cell Free Expression | | Descriptor: | 2,2',2''-NITRILOTRIETHANOL, Agmatine deiminase, CHLORIDE ION, ... | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-26 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Arabidopsis thaliana Agmatine Deiminase
To be Published
|
|
6BMG
 
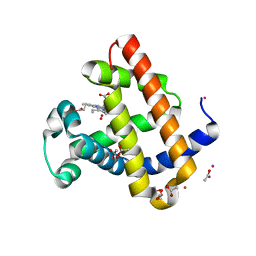 | | Structure of Recombinant Dwarf Sperm Whale Myoglobin (Oxy) | | Descriptor: | ACETATE ION, CADMIUM ION, Myoglobin, ... | | Authors: | Samuel, P.P, Miller, M.D, Xu, W, Alvarado, S, Phillips Jr, G.N, Olson, J.S. | | Deposit date: | 2017-11-14 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure of Recombinant Dwarf Sperm Whale Myoglobin (Oxy)
To Be Published
|
|
3CAZ
 
 | | Crystal structure of a BAR protein from Galdieria sulphuraria | | Descriptor: | BAR protein | | Authors: | McCoy, J.G, Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.344 Å) | | Cite: | Crystal structure of a BAR protein from Galdieria sulphuraria.
To be Published
|
|
4WKY
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmN KS2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-ketoacyl synthase, GLYCEROL, ... | | Authors: | Cuff, M.E, Mack, J.C, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-03 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5HOQ
 
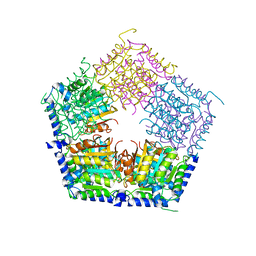 | | Apo structure of CalS11, TDP-rhamnose 3'-o-methyltransferase, an enzyme in Calicheamicin biosynthesis | | Descriptor: | SULFATE ION, TDP-rhamnose 3'-O-methyltransferase (CalS11) | | Authors: | Han, L, Helmich, K.E, Singh, S, Thorson, J.S, Bingman, C.A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Loop dynamics of thymidine diphosphate-rhamnose 3'-O-methyltransferase (CalS11), an enzyme in calicheamicin biosynthesis.
Struct Dyn., 3, 2016
|
|
3C9Q
 
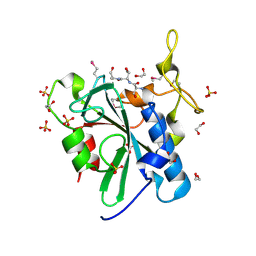 | | Crystal structure of the uncharacterized human protein C8orf32 with bound peptide | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, SULFATE ION, ... | | Authors: | Bitto, E, Bingman, C.A, McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-02-18 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the uncharacterized human protein C8orf32 with bound peptide.
To be Published
|
|
3IHR
 
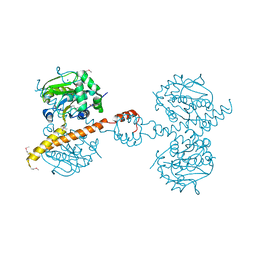 | | Crystal Structure of Uch37 | | Descriptor: | FORMIC ACID, SODIUM ION, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-11 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural characterization of human Uch37.
Proteins, 80, 2012
|
|
4XAU
 
 | | Crystal structure of AtS13 from Actinomadura melliaura | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative aminotransferase | | Authors: | Wang, F, Singh, S, Xu, W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-12-15 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.0012 Å) | | Cite: | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
4XRR
 
 | | Crystal structure of cals8 from micromonospora echinospora (P294S mutant) | | Descriptor: | CalS8, GLYCEROL | | Authors: | Michalska, K, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Characterization of CalS8, a TDP-alpha-D-Glucose Dehydrogenase Involved in Calicheamicin Aminodideoxypentose Biosynthesis.
J. Biol. Chem., 290, 2015
|
|
4XT0
 
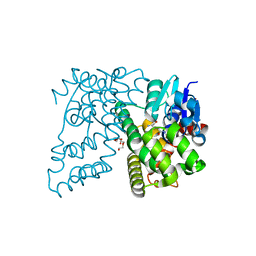 | | Crystal Structure of Beta-etherase LigF from Sphingobium sp. strain SYK-6 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTATHIONE, PENTAETHYLENE GLYCOL, ... | | Authors: | Helmich, K.E, Bingman, C.A, Donohue, T.J, Phillips Jr, G.N. | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis of Stereospecificity in the Bacterial Enzymatic Cleavage of beta-Aryl Ether Bonds in Lignin.
J.Biol.Chem., 291, 2016
|
|
5INJ
 
 | | Crystal Structure of Prenyltransferase PriB Ternary Complex with L-Tryptophan and Dimethylallyl thiolodiphosphate (DMSPP) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Prenyltransferase, ... | | Authors: | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-03-07 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
5MG1
 
 | | Structure of the photosensory module of Deinococcus phytochrome by serial femtosecond X-ray crystallography | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Young, I.D, Brewster, A.S, Clinger, J, Andi, B, Stan, C, Allaire, M, Nelsen, S, Alonso-Mori, R, Phillips Jr, G.N, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
3H7K
 
 | |
3GAN
 
 | | Crystal structure of gene product from Arabidopsis thaliana At3g22680 with bound suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, CHLORIDE ION, Uncharacterized protein At3g22680 | | Authors: | Burgie, E.S, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-02-17 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of gene product from Arabidopsis thaliana At3g22680 with bound suramin
To be Published
|
|
3TSR
 
 | | X-ray structure of mouse ribonuclease inhibitor complexed with mouse ribonuclease 1 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ribonuclease inhibitor, ... | | Authors: | Chang, A, Lomax, J.E, Bingman, C.A, Raines, R.T, Phillips Jr, G.N. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1999 Å) | | Cite: | Functional evolution of ribonuclease inhibitor: insights from birds and reptiles.
J.Mol.Biol., 426, 2014
|
|
3DHG
 
 | | Crystal Structure of Toluene 4-Monoxygenase Hydroxylase | | Descriptor: | AZIDE ION, CALCIUM ION, FE (III) ION, ... | | Authors: | Bailey, L.J, Mccoy, J.G, Phillips Jr, G.N, Fox, B.G. | | Deposit date: | 2008-06-17 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural consequences of effector protein complex formation in a diiron hydroxylase.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3G5T
 
 | | Crystal structure of trans-aconitate 3-methyltransferase from yeast | | Descriptor: | (2E)-2-(2-methoxy-2-oxoethyl)but-2-enedioic acid, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Burgie, E.S, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.119 Å) | | Cite: | Crystal structure of trans-aconitate 3-methyltransferase from yeast
To be Published
|
|
1TES
 
 | | OXYGEN BINDING MUSCLE PROTEIN | | Descriptor: | METHYLETHYLAMINE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Phillips Jr, G.N, Olson, J.S. | | Deposit date: | 1996-05-06 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of NO-induced oxidation of myoglobin and hemoglobin.
Biochemistry, 35, 1996
|
|
