1Q4R
 
 | | Gene Product of At3g17210 from Arabidopsis Thaliana | | Descriptor: | MAGNESIUM ION, protein At3g17210 | | Authors: | Phillips Jr, G.N, Bingman, C.A, Johnson, K.A, Smith, D.W, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-04 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the protein from gene At3g17210 of Arabidopsis thaliana
Proteins, 57, 2004
|
|
1Q44
 
 | | Crystal Structure of an Arabidopsis Thaliana Putative Steroid Sulfotransferase | | Descriptor: | MALONIC ACID, Steroid Sulfotransferase | | Authors: | Phillips Jr, G.N, Smith, D.W, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of At2g03760, a putative steroid sulfotransferase from Arabidopsis thaliana
Proteins, 57, 2004
|
|
1Q45
 
 | | 12-0xo-phytodienoate reductase isoform 3 | | Descriptor: | 12-oxophytodienoate-10,11-reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Smith, D.W, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of Arabidopsis At2g06050, 12-oxophytodienoate reductase isoform 3
Proteins, 58, 2005
|
|
1JW8
 
 | | 1.3 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF P6 FORM OF MYOGLOBIN | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Phillips Jr, G.N. | | Deposit date: | 2001-09-03 | | Release date: | 2001-10-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Sampling of the native conformational ensemble of myoglobin via structures in different crystalline environments.
Proteins, 70, 2008
|
|
2OHB
 
 | | Myoglobin cavity mutant I107W | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N, Schweers, R.L, Soman, J, Olson, J.S. | | Deposit date: | 2007-01-09 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand pathways in myoglobin: A review of trp cavity mutations.
Iubmb Life, 59, 2007
|
|
2OH9
 
 | | Myoglobin cavity mutant V68W | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N, Li, T, Brucker, E.A, Soman, J, Olson, J.S. | | Deposit date: | 2007-01-09 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand pathways in myoglobin: A review of trp cavity mutations.
Iubmb Life, 59, 2007
|
|
2OH8
 
 | | Myoglobin cavity mutant I28W | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N, Soman, J, Olson, J.S. | | Deposit date: | 2007-01-09 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand pathways in myoglobin: A review of trp cavity mutations.
Iubmb Life, 59, 2007
|
|
2OHA
 
 | | Myoglobin cavity mutant F138W | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N, Soman, J, Olson, J.S. | | Deposit date: | 2007-01-09 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand pathways in myoglobin: A review of trp cavity mutations.
Iubmb Life, 59, 2007
|
|
1LUE
 
 | | RECOMBINANT SPERM WHALE MYOGLOBIN H64D/V68A/D122N MUTANT (MET) | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N. | | Deposit date: | 2002-05-22 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular engineering of myoglobin: influence of residue 68 on the rate and the
enantioselectivity of oxidation reactions catalyzed by H64D/V68X myoglobin
Biochemistry, 42, 2003
|
|
1O16
 
 | | RECOMBINANT SPERM WHALE MYOGLOBIN H64D/V68S/D122N MUTANT (MET) | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N. | | Deposit date: | 2002-10-25 | | Release date: | 2003-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular engineering of myoglobin: influence of residue 68 on the rate and the
enantioselectivity of oxidation reactions catalyzed by H64D/V68X myoglobin
Biochemistry, 42, 2003
|
|
2GNX
 
 | | X-ray structure of a hypothetical protein from Mouse Mm.209172 | | Descriptor: | hypothetical protein | | Authors: | Phillips Jr, G.N, McCoy, J.G, Bitto, E, Wesenberg, G.E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-11 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | X-ray structure of a hypothetical protein from Mouse Mm.209172
To be Published
|
|
2TMA
 
 | |
4RNL
 
 | | The crystal structure of a possible galactose mutarotase from Streptomyces platensis subsp. rosaceus | | Descriptor: | GLYCEROL, PHOSPHATE ION, possible galactose mutarotase | | Authors: | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a possible galactose mutarotase from Streptomyces platensis subsp. rosaceus
To be Published
|
|
3KEV
 
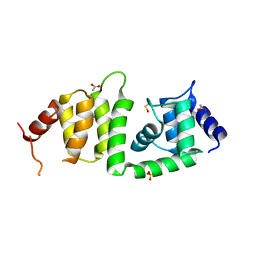 | | X-ray crystal structure of a DCUN1 domain-containing protein from Galdieria sulfuraria | | Descriptor: | ACETATE ION, Galieria sulfuraria DCUN1 domain-containing protein, SULFATE ION | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural architecture of Galdieria sulphuraria DCN1L.
Proteins, 79, 2011
|
|
6XOD
 
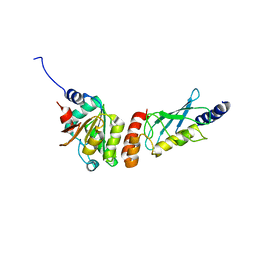 | | Crystal structure of the PEX4-PEX22 protein complex from Arabidopsis thaliana | | Descriptor: | Peroxisome biogenesis protein 22, Protein PEROXIN-4 | | Authors: | Olmos Jr, J.L, Bradford, S.E, Miller, M.D, Xu, W, Wright, Z.J, Bartel, B, Phillips Jr, G.N. | | Deposit date: | 2020-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Structure of the Arabidopsis PEX4-PEX22 Peroxin Complex-Insights Into Ubiquitination at the Peroxisomal Membrane
Front Cell Dev Biol, 10, 2022
|
|
6UVQ
 
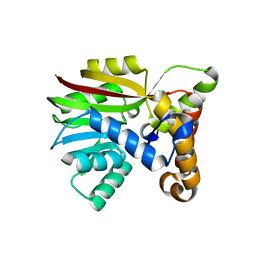 | | Crystal structure of Apo AtmM | | Descriptor: | ACETATE ION, D-glucose O-methyltransferase, MAGNESIUM ION | | Authors: | Alvarado, S.K, Wang, Z, Miller, M.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-11-04 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of Apo AtmM
To Be Published
|
|
6UWD
 
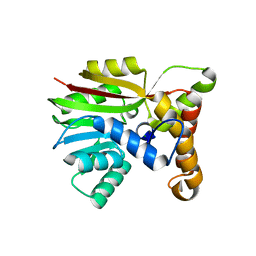 | | Crystal structure of Apo AtmM | | Descriptor: | ACETATE ION, D-glucose O-methyltransferase, MAGNESIUM ION | | Authors: | Alvarado, S.K, Wang, Z, Miller, M.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-11-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of Apo AtmM
To Be Published
|
|
6UPP
 
 | | Radiation Damage Test of PixJ Pb state crystals | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Clinger, J.A, Miller, M.D, Burgie, E.S, Vierstra, R.D, Phillips Jr, G.N. | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UV6
 
 | | AtmM with bound rebeccamycin analogue | | Descriptor: | 12-beta-D-glucopyranosyl-12,13-dihydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazole-5,7(6H)-dione, D-glucose O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Alvarado, S.K, Wang, Z, Miller, M.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure of AtmM Bound with Glycosylated Indolocarbazole
To Be Published
|
|
6V04
 
 | | DynU16 crystal structure, a putative protein in the dynemicin biosynthetic locus | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Alvarado, S.K, Miller, M.D, Bhardwaj, M, Thorson, J.S, Van Lanen, S.G, Phillips Jr, G.N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of DynU16, a START/Bet v1-like protein involved in dynemicin biosynthesis.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
5HOQ
 
 | | Apo structure of CalS11, TDP-rhamnose 3'-o-methyltransferase, an enzyme in Calicheamicin biosynthesis | | Descriptor: | SULFATE ION, TDP-rhamnose 3'-O-methyltransferase (CalS11) | | Authors: | Han, L, Helmich, K.E, Singh, S, Thorson, J.S, Bingman, C.A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Loop dynamics of thymidine diphosphate-rhamnose 3'-O-methyltransferase (CalS11), an enzyme in calicheamicin biosynthesis.
Struct Dyn., 3, 2016
|
|
5INJ
 
 | | Crystal Structure of Prenyltransferase PriB Ternary Complex with L-Tryptophan and Dimethylallyl thiolodiphosphate (DMSPP) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Prenyltransferase, ... | | Authors: | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-03-07 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
7ML6
 
 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | Descriptor: | CalU17, GLYCEROL | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2021-04-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
6P58
 
 | | Dark and Steady State-Illuminated Crystal Structure of Cyanobacteriochrome Receptor PixJ at 150K | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Clinger, J.A, Miller, M.D, Buirgie, E.S, Vierstra, R.D, Phillips Jr, G.N. | | Deposit date: | 2019-05-29 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P9V
 
 | | Crystal Structure of hMAT Mutant K289L | | Descriptor: | ADENOSINE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Miller, M.D, Xu, W, Huber, T.D, Clinger, J.A, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Methionine Adenosyltransferase Engineering to Enable Bioorthogonal Platforms for AdoMet-Utilizing Enzymes.
Acs Chem.Biol., 15, 2020
|
|
