2AB4
 
 | |
1ZE1
 
 | |
1ZE2
 
 | |
6B55
 
 | | Crystal structure of the Plant Defensin NaD1 complexed with phosphatidic acid | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, 1,2-ETHANEDIOL, Flower-specific defensin, ... | | Authors: | Jarva, M, Phan, K, Humble, C, Lay, F.T, Hulett, M, Kvansakul, M. | | Deposit date: | 2017-09-28 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of a carpet-like antimicrobial defensin-phospholipid membrane disruption complex.
Nat Commun, 9, 2018
|
|
6CS9
 
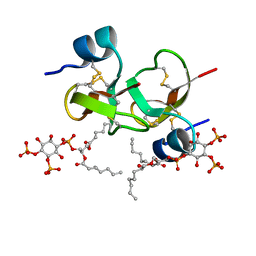 | | Crystal structure of human beta-defensin 2 in complex with PIP2 | | Descriptor: | Beta-defensin 4A, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Jarva, M, Phan, K, Lay, F.T, Humble, C, Hulett, M, Kvansakul, M. | | Deposit date: | 2018-03-20 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Human beta-defensin 2 killsCandida albicansthrough phosphatidylinositol 4,5-bisphosphate-mediated membrane permeabilization.
Sci Adv, 4, 2018
|
|
7JUK
 
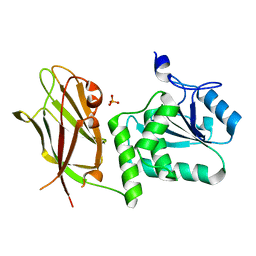 | | Crystal structure of PTEN with a tetra-phosphorylated tail (4p-crPTEN-13sp-T2, SDTTDSDPENEG) | | Descriptor: | PHOSPHATE ION, Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | | Authors: | Dempsey, D, Phan, K, Cole, P, Gabelli, S.B. | | Deposit date: | 2020-08-19 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The structural basis of PTEN regulation by multi-site phosphorylation.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7JVX
 
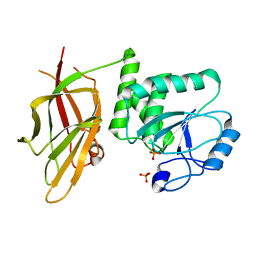 | | Crystal structure of PTEN (aa 7-353 followed by spacer TGGGSGGTGGGSGGTGGGCY ligated to peptide pSDpTpTDpSDPENEPFDED) | | Descriptor: | PHOSPHATE ION, Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | | Authors: | Dempsey, D, Phan, K, Cole, P, Gabelli, S.B. | | Deposit date: | 2020-08-24 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis of PTEN regulation by multi-site phosphorylation.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7JUL
 
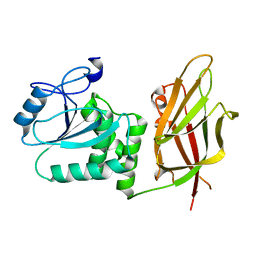 | | Crystal structure of non phosphorylated PTEN (n-crPTEN-13sp-T1, SDTTDSDPENEG) | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | | Authors: | Dempsey, D, Phan, K, Cole, P, Gabelli, S.B. | | Deposit date: | 2020-08-20 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The structural basis of PTEN regulation by multi-site phosphorylation.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KU0
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 138 (yellow) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU1
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 139 (green) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU2
 
 | | Data clustering and dynamics of chymotrypsinogen clulster 140 (structure) | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU3
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 141 (cyan) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTZ
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 131 (purple) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTY
 
 | | Data clustering and dynamics of chymotrypsinogen average structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Shi, W, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
