1FBR
 
 | |
8FBP
 
 | | Glutamine synthetase from Pseudomonas aeruginosa, filament double-unit in compressed conformation | | Descriptor: | Glutamine synthetase | | Authors: | Phan, I.Q, Staker, B, Shek, R, Moser, T.H, Evans, J.E, van Voorhis, W.C, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-11-29 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Glutamine synthetase from Pseudomonas aeruginosa, filament double-unit in compressed conformation
To Be Published
|
|
1OIG
 
 | | The solution structure of the DPY module from the Dumpy protein | | Descriptor: | Dumpy, isoform Y | | Authors: | Wilkin, M.B, Becker, M.N, Mulvey, D, Phan, I, Chao, A, Cooper, K, Chung, H.J, Campbell, I.D, Baron, M, MacIntyre, R. | | Deposit date: | 2003-06-18 | | Release date: | 2003-06-26 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Drosophila Dumpy is a Gigantic Extracellular Protein Required to Maintain Tension at Epidermal-Cuticle Attachment Sites
Curr.Biol., 10, 2000
|
|
8FBQ
 
 | | Crystal structure of Plasmodium vivax glycylpeptide N-tetradecanoyltransferase (N-myristoyltransferase, NMT) bound to myristoyl-CoA and inhibitor 12b | | Descriptor: | 1-[(3M)-3-{3-[2-(1,3,5-trimethyl-1H-pyrazol-4-yl)ethoxy]pyridin-2-yl}phenyl]piperazine, ACETATE ION, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Staker, B.L, Lovell, S.W, Phan, I.Q, Early, J, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-11-29 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of potent and selective N-myristoyltransferase inhibitors of Plasmodium vivax liver stage hypnozoites and schizonts.
Nat Commun, 14, 2023
|
|
3KJR
 
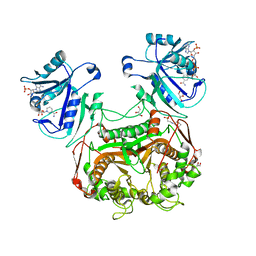 | | Crystal structure of dihydrofolate reductase/thymidylate synthase from Babesia bovis determined using SlipChip based microfluidics | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Dihydrofolate reductase/thymidylate synthase, GLYCEROL, ... | | Authors: | Li, L, Du, W, Edwards, T.E, Staker, B.L, Phan, I, Stacy, R, Ismagilov, R.F, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-11-03 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Multiparameter screening on SlipChip used for nanoliter protein crystallization combining free interface diffusion and microbatch methods.
J.Am.Chem.Soc., 132, 2010
|
|
8VKA
 
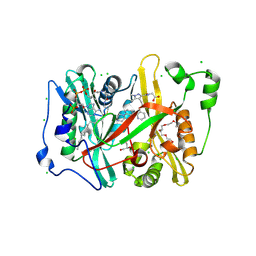 | | Crystal structure of Plasmodium vivax glycylpeptide N-tetradecanoyltransferase (N-myristoyltransferase, NMT) bound to myristoyl-CoA and inhibitor 9c | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Fenwick, M.K, Staker, B.L, Phan, I.Q, Early, J, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Exploring Subsite Selectivity within Plasmodium vivax N -Myristoyltransferase Using Pyrazole-Derived Inhibitors.
J.Med.Chem., 67, 2024
|
|
8VKB
 
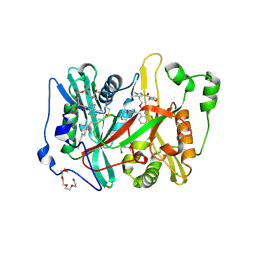 | | Crystal structure of Plasmodium vivax glycylpeptide N-tetradecanoyltransferase (N-myristoyltransferase, NMT) bound to myristoyl-CoA and inhibitor 10b | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase, ... | | Authors: | Fenwick, M.K, Staker, B.L, Phan, I.Q, Early, J, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Exploring Subsite Selectivity within Plasmodium vivax N -Myristoyltransferase Using Pyrazole-Derived Inhibitors.
J.Med.Chem., 67, 2024
|
|
3IFT
 
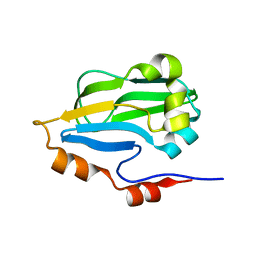 | | Crystal structure of glycine cleavage system protein H from Mycobacterium tuberculosis, using X-rays from the Compact Light Source. | | Descriptor: | Glycine cleavage system H protein | | Authors: | Edwards, T.E, Abendroth, J, Staker, B, Mayer, C, Phan, I, Kelley, A, Analau, E, Leibly, D, Rifkin, J, Loewen, R, Ruth, R.D, Stewart, L.J, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2009-07-25 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure determination of the glycine cleavage system protein H of Mycobacterium tuberculosis using an inverse Compton synchrotron X-ray source.
J.Struct.Funct.Genom., 11, 2010
|
|
1DS6
 
 | | CRYSTAL STRUCTURE OF A RAC-RHOGDI COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAS-RELATED C3 BOTULINUM TOXIN SUBSTRATE 2, ... | | Authors: | Scheffzek, K, Stephan, I, Jensen, O.N, Illenberger, D, Gierschik, P. | | Deposit date: | 2000-01-07 | | Release date: | 2000-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Rac-RhoGDI complex and the structural basis for the regulation of Rho proteins by RhoGDI.
Nat.Struct.Biol., 7, 2000
|
|
