1PVN
 
 | | The crystal structure of the complex between IMP dehydrogenase catalytic domain and a transition state analogue MZP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-CARBAMOYL-1-BETA-D-RIBOFURANOSYL-IMIDAZOLIUM-5-OLATE-5'-PHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Gan, L, Seyedsayamdost, M, Shuto, S, Matsuda, A, Petsko, G.A, Hedstrom, L. | | Deposit date: | 2003-06-27 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Immunosuppressive Agent Mizoribine Monophosphate Forms a Transition State Analogue Complex with Inosine Monophosphate Dehydrogenase
Biochemistry, 42, 2003
|
|
1RTQ
 
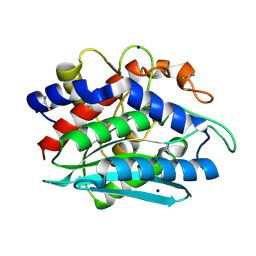 | | The 0.95 Angstrom Resolution Crystal Structure of the Aminopeptidase from Aeromonas proteolytica | | Descriptor: | Bacterial leucyl aminopeptidase, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Desmarais, W, Bienvenue, D.L, Krzysztof, B.P, Holz, R.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2003-12-10 | | Release date: | 2004-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The high-resolution structures of the neutral and the low pH crystals of aminopeptidase from Aeromonas proteolytica.
J.Biol.Inorg.Chem., 11, 2006
|
|
1Q0X
 
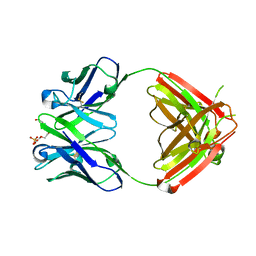 | | Anti-morphine Antibody 9B1 Unliganded Form | | Descriptor: | Fab 9B1, heavy chain, light chain, ... | | Authors: | Pozharski, E, Wilson, M.A, Hewagama, A, Shanafelt, A.B, Petsko, G, Ringe, D. | | Deposit date: | 2003-07-17 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Anchoring a cationic ligand: the structure of the Fab fragment of the anti-morphine antibody 9B1 and its complex with morphine
J.Mol.Biol., 337, 2004
|
|
1S5M
 
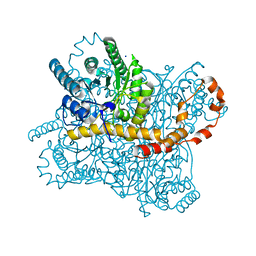 | | Xylose Isomerase in Substrate and Inhibitor Michaelis States: Atomic Resolution Studies of a Metal-Mediated Hydride Shift | | Descriptor: | MANGANESE (II) ION, SODIUM ION, Xylose isomerase, ... | | Authors: | Fenn, T.D, Ringe, D, Petsko, G.A. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Xylose isomerase in substrate and inhibitor michaelis States: atomic resolution studies of a metal-mediated hydride shift(,).
Biochemistry, 43, 2004
|
|
1ELC
 
 | | Analogous inhibitors of elastase do not always bind analogously | | Descriptor: | 6-ammonio-N-(trifluoroacetyl)-L-norleucyl-N-[4-(1-methylethyl)phenyl]-L-phenylalaninamide, CALCIUM ION, ELASTASE | | Authors: | Mattos, C, Rasmussen, B, Ding, X, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-04-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Analogous inhibitors of elastase do not always bind analogously.
Nat.Struct.Biol., 1, 1994
|
|
1ELA
 
 | | Analogous inhibitors of elastase do not always bind analogously | | Descriptor: | 6-ammonio-N-(trifluoroacetyl)-L-norleucyl-N-[4-(1-methylethyl)phenyl]-L-prolinamide, ACETIC ACID, CALCIUM ION, ... | | Authors: | Mattos, C, Rasmussen, B, Ding, X, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-04-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analogous inhibitors of elastase do not always bind analogously.
Nat.Struct.Biol., 1, 1994
|
|
1ELB
 
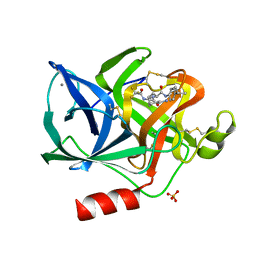 | | Analogous inhibitors of elastase do not always bind analogously | | Descriptor: | 6-ammonio-N-(trifluoroacetyl)-L-norleucyl-N-[4-(1-methylethyl)phenyl]-L-leucinamide, CALCIUM ION, ELASTASE, ... | | Authors: | Mattos, C, Rasmussen, B, Ding, X, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-06-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analogous inhibitors of elastase do not always bind analogously.
Nat.Struct.Biol., 1, 1994
|
|
1BKH
 
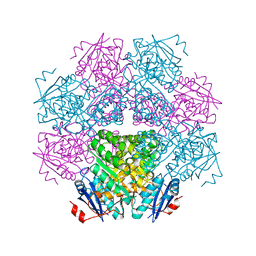 | | MUCONATE LACTONIZING ENZYME FROM PSEUDOMONAS PUTIDA | | Descriptor: | MUCONATE LACTONIZING ENZYME | | Authors: | Hasson, M.S, Schlichting, I, Moulai, J, Taylor, K, Barrett, W, Kenyon, G.L, Babbitt, P.C, Gerlt, J.A, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-07-07 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of an enzyme active site: the structure of a new crystal form of muconate lactonizing enzyme compared with mandelate racemase and enolase.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BRM
 
 | | ASPARTATE BETA-SEMIALDEHYDE DEHYDROGENASE FROM ESCHERICHIA COLI | | Descriptor: | ASPARTATE-SEMIALDEHYDE DEHYDROGENASE | | Authors: | Hadfield, A.T, Kryger, G, Ouyang, J, Ringe, D, Petsko, G.A, Viola, R.E. | | Deposit date: | 1998-08-24 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of aspartate-beta-semialdehyde dehydrogenase from Escherichia coli, a key enzyme in the aspartate family of amino acid biosynthesis.
J.Mol.Biol., 289, 1999
|
|
1QIS
 
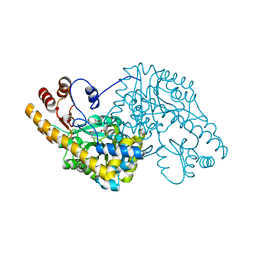 | | ASPARTATE AMINOTRANSFERASE FROM ESCHERICHIA COLI, C191F MUTATION, WITH BOUND MALEATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jeffery, C.J, Gloss, L.M, Petsko, G.A, Ringe, D. | | Deposit date: | 1999-06-15 | | Release date: | 2000-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Role of Residues Outside the Active Site in Catalysis: Structural Basis for Function of C191 Mutants of E. Coli Aspartate Aminotransferase
Protein Eng., 13, 2000
|
|
1QIT
 
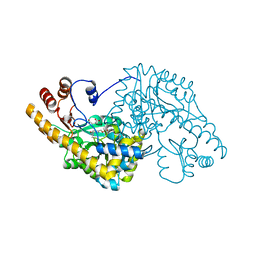 | | ASPARTATE AMINOTRANSFERASE FROM ESCHERICHIA COLI, C191W MUTATION, WITH BOUND MALEATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jeffery, C.J, Gloss, L.M, Petsko, G.A, Ringe, D. | | Deposit date: | 1999-06-15 | | Release date: | 2000-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Role of Residues Outside the Active Site in Catalysis: Structural Basis for Function of C191 Mutants of E. Coli Aspartate Aminotransferase
Protein Eng., 13, 2000
|
|
1RIU
 
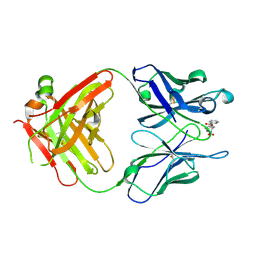 | | Anti-Cocaine Antibody M82G2 Complexed With Norbenzoylecgonine | | Descriptor: | 3-(BENZOYLOXY)-8-AZA-BICYCLO[3.2.1]OCTANE-2-CARBOXYLIC ACID, Fab M82G2, Heavy Chain, ... | | Authors: | Pozharski, E, Hewagama, A, Shanafelt, A, Petsko, G, Ringe, D. | | Deposit date: | 2003-11-18 | | Release date: | 2003-12-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carving a Binding Site: Structural Study of an Anti-Cocaine Antibody in Complex with Three Cocaine Analogs
To be Published
|
|
1RFD
 
 | | ANTI-COCAINE ANTIBODY M82G2 | | Descriptor: | Fab M82G2, Heavy Chain, Light Chain | | Authors: | Pozharski, E, Hewagama, A, Shanafelt, A.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2003-11-07 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Diversity in hapten recognition: structural study of an anti-cocaine antibody M82G2.
J.Mol.Biol., 349, 2005
|
|
1QIR
 
 | | ASPARTATE AMINOTRANSFERASE FROM ESCHERICHIA COLI, C191Y MUTATION, WITH BOUND MALEATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jeffery, C.J, Gloss, L.M, Petsko, G.A, Ringe, D. | | Deposit date: | 1999-06-15 | | Release date: | 2000-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Role of Residues Outside the Active Site in Catalysis: Structural Basis for Function of C191 Mutants of E. Coli Aspartate Aminotransferase
Protein Eng., 13, 2000
|
|
1Q72
 
 | | Anti-Cocaine Antibody M82G2 Complexed with Cocaine | | Descriptor: | COCAINE, Fab M82G2, Heavy chain, ... | | Authors: | Pozharski, E, Moulin, A, Hewagama, A, Shanafelt, A.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2003-08-15 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Diversity in hapten recognition: structural study of an anti-cocaine antibody M82G2.
J.Mol.Biol., 349, 2005
|
|
1EY3
 
 | |
1RAT
 
 | |
1ESA
 
 | | DIRECT STRUCTURE OBSERVATION OF AN ACYL-ENZYME INTERMEDIATE IN THE HYDROLYSIS OF AN ESTER SUBSTRATE BY ELASTASE | | Descriptor: | CALCIUM ION, PORCINE PANCREATIC ELASTASE, SULFATE ION | | Authors: | Ding, X, Rasmussen, B, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-04 | | Release date: | 1994-04-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct structural observation of an acyl-enzyme intermediate in the hydrolysis of an ester substrate by elastase.
Biochemistry, 33, 1994
|
|
1RIV
 
 | | Anti-Cocaine Antibody M82G2 Complexed With meta-Oxybenzoylecgonine | | Descriptor: | 3-(3-HYDROXY-BENZOYLOXY)-8-METHYL-8-AZA-BICYCLO[3.2.1]OCTANE-2-CARBOXYLIC ACID, Fab M82G2, Heavy Chain, ... | | Authors: | Pozharski, E, Hewagama, A, Shanafelt, A, Petsko, G, Ringe, D. | | Deposit date: | 2003-11-18 | | Release date: | 2003-12-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Carving a Binding Site: Structural Study of an Anti-Cocaine Antibody in Complex with Three Cocaine Analogs
To be Published
|
|
1B4X
 
 | | ASPARTATE AMINOTRANSFERASE FROM E. COLI, C191S MUTATION, WITH BOUND MALEATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jeffery, C.J, Gloss, L.M, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-12-30 | | Release date: | 2000-10-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The role of residues outside the active site: structural basis for function of C191 mutants of Escherichia coli aspartate aminotransferase.
Protein Eng., 13, 2000
|
|
1AZ2
 
 | | CITRATE BOUND, C298A/W219Y MUTANT HUMAN ALDOSE REDUCTASE | | Descriptor: | ALDOSE REDUCTASE, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H, Bohren, K.M, Ringe, D, Petsko, G.A, Gabbay, K.H. | | Deposit date: | 1997-11-24 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The alrestatin double-decker: binding of two inhibitor molecules to human aldose reductase reveals a new specificity determinant.
Biochemistry, 36, 1997
|
|
1DQR
 
 | | CRYSTAL STRUCTURE OF RABBIT PHOSPHOGLUCOSE ISOMERASE, A GLYCOLYTIC ENZYME THAT MOONLIGHTS AS NEUROLEUKIN, AUTOCRINE MOTILITY FACTOR, AND DIFFERENTIATION MEDIATOR | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, PHOSPHOGLUCOSE ISOMERASE | | Authors: | Bahnson, B.J, Jeffery, C.J, Ringe, D, Petsko, G.A. | | Deposit date: | 2000-01-05 | | Release date: | 2000-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of rabbit phosphoglucose isomerase, a glycolytic enzyme that moonlights as neuroleukin, autocrine motility factor, and differentiation mediator.
Biochemistry, 39, 2000
|
|
1AZ1
 
 | | ALRESTATIN BOUND TO C298A/W219Y MUTANT HUMAN ALDOSE REDUCTASE | | Descriptor: | ALDOSE REDUCTASE, ALRESTATIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Bohren, K.M, Petsko, G.A, Ringe, D, Gabbay, K.H. | | Deposit date: | 1997-11-24 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The alrestatin double-decker: binding of two inhibitor molecules to human aldose reductase reveals a new specificity determinant.
Biochemistry, 36, 1997
|
|
1CP6
 
 | | 1-BUTANEBORONIC ACID BINDING TO AEROMONAS PROTEOLYTICA AMINOPEPTIDASE | | Descriptor: | 1-BUTANE BORONIC ACID, PROTEIN (AMINOPEPTIDASE), ZINC ION | | Authors: | Depaola, C.C, Bennett, B, Holz, R.C, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-06-08 | | Release date: | 1999-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1-Butaneboronic acid binding to Aeromonas proteolytica aminopeptidase: a case of arrested development.
Biochemistry, 38, 1999
|
|
1EEP
 
 | | 2.4 A RESOLUTION CRYSTAL STRUCTURE OF BORRELIA BURGDORFERI INOSINE 5'-MONPHOSPHATE DEHYDROGENASE IN COMPLEX WITH A SULFATE ION | | Descriptor: | INOSINE 5'-MONOPHOSPHATE DEHYDROGENASE, SULFATE ION | | Authors: | McMillan, F.M, Cahoon, M, White, A, Hedstrom, L, Petsko, G.A, Ringe, D. | | Deposit date: | 2000-02-01 | | Release date: | 2000-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure at 2.4 A resolution of Borrelia burgdorferi inosine 5'-monophosphate dehydrogenase: evidence of a substrate-induced hinged-lid motion by loop 6.
Biochemistry, 39, 2000
|
|
