5T4L
 
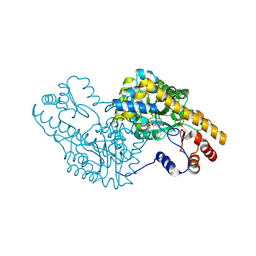 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | Descriptor: | (4R)-4-amino-6-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}hexanoic acid, Aspartate aminotransferase | | Authors: | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2016-08-29 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T4K
 
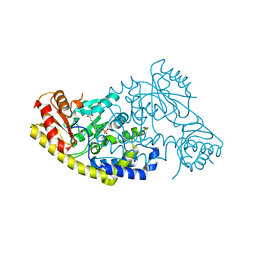 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | Descriptor: | (4S)-5-fluoro-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]pentanoic acid, HTH-type transcriptional regulatory protein GabR | | Authors: | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2016-08-29 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T4J
 
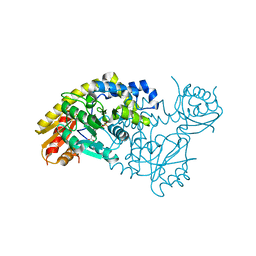 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, HTH-type transcriptional regulatory protein GabR, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2016-08-29 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UNO
 
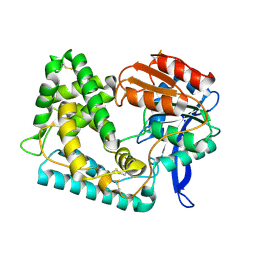 | | Crystal Structure of Hip1 (Rv2224c) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, White, A, Goldfarb, N, Milne, A.C, Liu, D, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2017-01-31 | | Release date: | 2017-04-12 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c).
Biochemistry, 56, 2017
|
|
5UOH
 
 | | Crystal Structure of Hip1 (Rv2224c) T466A mutant | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, White, A, Goldfarb, N, Milne, A.C, Liu, D, Baikovitz, J, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2017-01-31 | | Release date: | 2017-04-12 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c).
Biochemistry, 56, 2017
|
|
2CHF
 
 | | STRUCTURE OF THE MG2+-BOUND FORM OF CHEY AND THE MECHANISM OF PHOSPHORYL TRANSFER IN BACTERIAL CHEMOTAXIS | | Descriptor: | CHEY | | Authors: | Stock, A, Martinez-Hackert, E, Rasmussen, B, West, A, Stock, J, Ringe, D, Petsko, G. | | Deposit date: | 1994-01-17 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Mg(2+)-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis.
Biochemistry, 32, 1993
|
|
2CHE
 
 | | STRUCTURE OF THE MG2+-BOUND FORM OF CHEY AND MECHANISM OF PHOSPHORYL TRANSFER IN BACTERIAL CHEMOTAXIS | | Descriptor: | CHEY, MAGNESIUM ION | | Authors: | Stock, A, Martinez-Hackert, E, Rasmussen, B, West, A, Stock, J, Ringe, D, Petsko, G. | | Deposit date: | 1994-01-17 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Mg(2+)-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis.
Biochemistry, 32, 1993
|
|
3B3C
 
 | | Crystal structure of the M180A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine phosphonic acid | | Descriptor: | Bacterial leucyl aminopeptidase, LEUCINE PHOSPHONIC ACID, POTASSIUM ION, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-19 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
3B3S
 
 | | Crystal structure of the M180A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine | | Descriptor: | Bacterial leucyl aminopeptidase, LEUCINE, SODIUM ION, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-22 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
4QXW
 
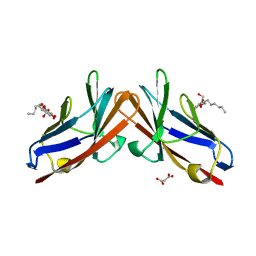 | | Crystal structure of the human CEACAM1 membrane distal amino terminal (N)-domain | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, MALONIC ACID, octyl beta-D-glucopyranoside | | Authors: | Huang, Y.H, Gandhi, A.K, Russell, A, Kondo, Y, Chen, Q, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2014-07-22 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | CEACAM1 regulates TIM-3-mediated tolerance and exhaustion.
Nature, 517, 2015
|
|
3B3V
 
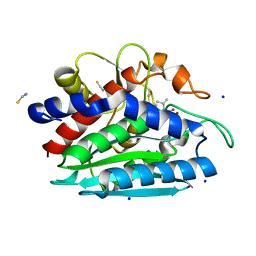 | | Crystal structure of the S228A mutant of the aminopeptidase from Vibrio proteolyticus | | Descriptor: | Bacterial leucyl aminopeptidase, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Zahniser, M.P.D, Milne, A, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-22 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
1TIM
 
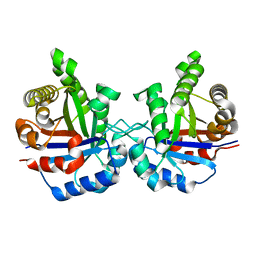 | | STRUCTURE OF TRIOSE PHOSPHATE ISOMERASE FROM CHICKEN MUSCLE | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Banner, D.W, Bloomer, A.C, Petsko, G.A, Phillips, D.C, Wilson, I.A. | | Deposit date: | 1976-09-01 | | Release date: | 1976-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic coordinates for triose phosphate isomerase from chicken muscle.
Biochem.Biophys.Res.Commun., 72, 1976
|
|
2QB3
 
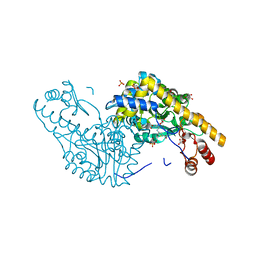 | | Structural Studies Reveal the Inactivation of E. coli L-Aspartate Aminotransferase by (s)-4,5-dihydro-2-thiophenecarboxylic acid (SADTA) via Two Mechanisms (at pH 7.5) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Lepore, B, Fu, M, Silverman, R.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-06-15 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Inactivation of Escherichia coli L-aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-thiophenecarboxylic acid reveals "a tale of two mechanisms".
Biochemistry, 46, 2007
|
|
2QBT
 
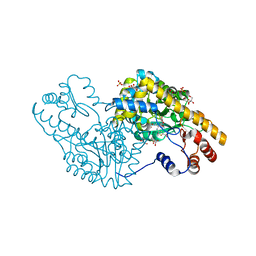 | | Structural Studies Reveal The Inactivation of E. coli L-aspartate aminotransferase by (S)-4,5-amino-dihydro-2-thiophenecarboxylic acid (SADTA) via Two Mechanisms (at pH 8.0) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Lepore, B, Fu, M, Silverman, R.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-06-18 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inactivation of Escherichia coli l-Aspartate Aminotransferase by (S)-4-Amino-4,5-dihydro-2-thiophenecarboxylic Acid Reveals "A Tale of Two Mechanisms".
Biochemistry, 46, 2007
|
|
2QA3
 
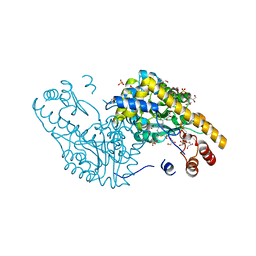 | | Structural Studies Reveal the Inactivation of E. coli L-aspartate aminotransferase by (S)-4,5-amino-dihydro-2-thiophenecarboxylic acid (SADTA) via two mechanisms (at pH6.5) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Lepore, B, Fu, M, Silverman, R.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-06-14 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inactivation of Escherichia coli L-aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-thiophenecarboxylic acid reveals "a tale of two mechanisms".
Biochemistry, 46, 2007
|
|
2R2D
 
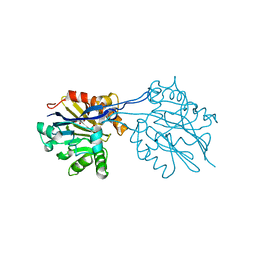 | | Structure of a quorum-quenching lactonase (AiiB) from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, PHOSPHATE ION, ZINC ION, ... | | Authors: | Liu, D, Thomas, P.W, Momb, J, Hoang, Q, Petsko, G.A, Ringe, D, Fast, W. | | Deposit date: | 2007-08-24 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and specificity of a quorum-quenching lactonase (AiiB) from Agrobacterium tumefaciens.
Biochemistry, 46, 2007
|
|
2Q7W
 
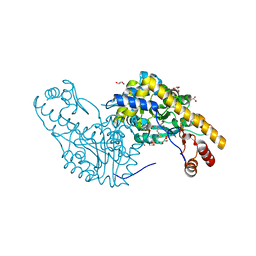 | | Structural Studies Reveals the Inactivation of E. coli L-aspartate aminotransferase (S)-4,5-amino-dihydro-2-thiophenecarboxylic acid (SADTA) via two mechanisms at pH 6.0 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Lepore, B, Fu, M, Silverman, R.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-06-07 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inactivation of Escherichia coli l-Aspartate Aminotransferase by (S)-4-Amino-4,5-dihydro-2-thiophenecarboxylic Acid Reveals "A Tale of Two Mechanisms".
Biochemistry, 46, 2007
|
|
2QB2
 
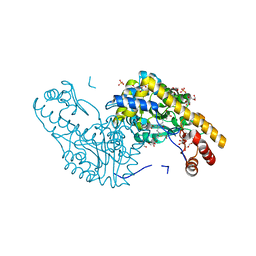 | | Structural Studies Reveal the Inactivation of E. coli L-aspartate aminotransferase by (s)-4,5-dihydro-2thiophenecarboylic acid (SADTA) via two mechanisms (at pH 7.0). | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Lepore, B, Fu, M, Silverman, R.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-06-15 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inactivation of Escherichia coli L-aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-thiophenecarboxylic acid reveals "a tale of two mechanisms".
Biochemistry, 46, 2007
|
|
1DTN
 
 | |
3RHN
 
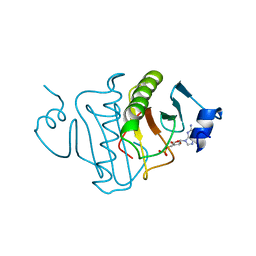 | | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN (HINT) FROM RABBIT COMPLEXED WITH GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN | | Authors: | Brenner, C, Garrison, P, Gilmour, J, Peisach, D, Ringe, D, Petsko, G.A, Lowenstein, J.M. | | Deposit date: | 1997-02-11 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins.
Nat.Struct.Biol., 4, 1997
|
|
2BBG
 
 | |
3RAT
 
 | |
1MBC
 
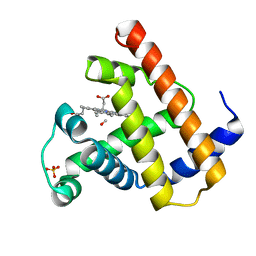 | |
1MDR
 
 | | THE ROLE OF LYSINE 166 IN THE MECHANISM OF MANDELATE RACEMASE FROM PSEUDOMONAS PUTIDA: MECHANISTIC AND CRYSTALLOGRAPHIC EVIDENCE FOR STEREOSPECIFIC ALKYLATION BY (R)-ALPHA-PHENYLGLYCIDATE | | Descriptor: | ATROLACTIC ACID (2-PHENYL-LACTIC ACID), MAGNESIUM ION, MANDELATE RACEMASE | | Authors: | Landro, J.A, Gerlt, J.A, Kozarich, J.W, Koo, C.W, Shah, V.J, Kenyon, G.L, Neidhart, D.J, Fujita, S, Petsko, G.A. | | Deposit date: | 1993-11-19 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of lysine 166 in the mechanism of mandelate racemase from Pseudomonas putida: mechanistic and crystallographic evidence for stereospecific alkylation by (R)-alpha-phenylglycidate.
Biochemistry, 33, 1994
|
|
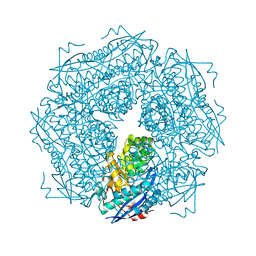
2FOF
 
 | | Structure of porcine pancreatic elastase in 80% isopropanol | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Mattos, C, Bellamacina, C.R, Peisach, E, Pereira, A, Vitkup, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple solvent crystal structures: Probing binding sites, plasticity and hydration
J.Mol.Biol., 357, 2006
|
|
