8PD9
 
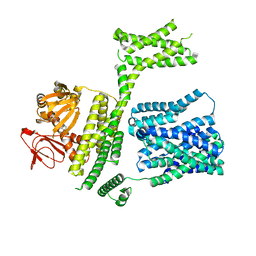 | | cAMP-bound SpSLC9C1 in lipid nanodiscs, protomer state 1 | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Sperm-specific sodium proton exchanger | | Authors: | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | Deposit date: | 2023-06-12 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
8PDV
 
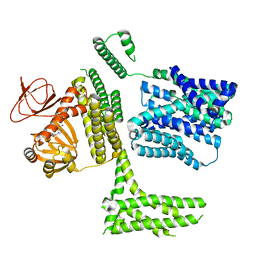 | | cGMP-bound SpSLC9C1 in lipid nanodiscs, protomer | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Sperm-specific sodium proton exchanger | | Authors: | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | Deposit date: | 2023-06-12 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
8PDU
 
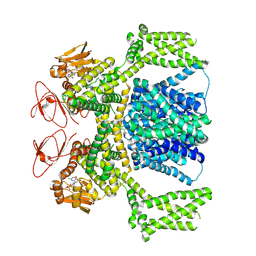 | | cGMP-bound SpSLC9C1 in lipid nanodiscs, dimer | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Sperm-specific sodium proton exchanger | | Authors: | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | Deposit date: | 2023-06-12 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
1UR8
 
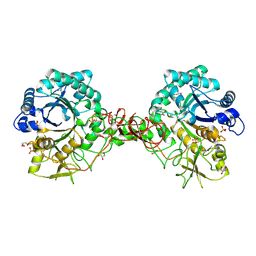 | | Interactions of a family 18 chitinase with the designed inhibitor HM508, and its degradation product, chitobiono-delta-lactone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, CHITINASE B, GLYCEROL, ... | | Authors: | Vaaje-Kolstad, G, Vasella, A, Peter, M.G, Netter, C, Houston, D.R, Westereng, B, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2003-10-27 | | Release date: | 2004-04-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of a Family 18 Chitinase with the Designed Inhibitor Hm508 and its Degradation Product, Chitobiono-Delta-Lactone.
J.Biol.Chem., 279, 2004
|
|
1UR9
 
 | | Interactions of a family 18 chitinase with the designed inhibitor HM508, and its degradation product, chitobiono-delta-lactone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, CHITINASE B, GLYCEROL, ... | | Authors: | Vaaje-Kolstad, G, Vasella, A, Peter, M.G, Netter, C, Houston, D.R, Westereng, B, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2003-10-27 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interactions of a Family 18 Chitinase with the Designed Inhibitor Hm508 and its Degradation Product, Chitobiono-Delta-Lactone.
J.Biol.Chem., 279, 2004
|
|
1OGB
 
 | | Chitinase b from Serratia marcescens mutant D142N | | Descriptor: | CHITINASE B, GLYCEROL, SULFATE ION | | Authors: | Vaaje-Kolstad, G, Houston, D.R, Rao, F.V, Peter, M.G, Synstad, B, van Aalten, D.M.F, Eijsink, V.G.H. | | Deposit date: | 2003-04-29 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the D142N Mutant of the Family 18 Chitinase Chib from Serratia Marcescens and its Complex with Allosamidin
Biochim.Biophys.Acta, 1696, 2004
|
|
1H0G
 
 | | Complex of a chitinase with the natural product cyclopentapeptide argadin from Clonostachys | | Descriptor: | Argadin, CHITINASE B, GLYCEROL | | Authors: | Houston, D, Shiomi, K, Arai, N, Omura, S, Peter, M.G, Turberg, A, Synstad, B, Eijsink, V.G.H, Aalten, D.M.F. | | Deposit date: | 2002-06-19 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Inhibited Complexes of a Chitinase with Natural Product Cyclopentapeptides - Peptide Mimicry of a Carbohydrate Substrate
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1H0I
 
 | | Complex of a chitinase with the natural product cyclopentapeptide argifin from Gliocladium | | Descriptor: | ARGIFIN, CHITINASE B, GLYCEROL, ... | | Authors: | Houston, D.R, Shiomi, K, Arai, N, Omura, S, Peter, M.G, Turberg, A, Synstad, B, Eijsink, V.G.H, Aalten, D.M.F. | | Deposit date: | 2002-06-19 | | Release date: | 2002-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Inhibited Complexes of a Chitinase with Natural Product Cyclopentapeptides - Peptide Mimicry of a Carbohydrate Substrate
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
8CP7
 
 | |
2L4F
 
 | |
2L4E
 
 | |
2KWU
 
 | | Solution Structure of UBM2 of murine Polymerase iota in Complex with Ubiquitin | | Descriptor: | DNA polymerase iota, Ubiquitin | | Authors: | Burschowsky, D, Rudolf, F, Rabut, G, Herrmann, T, Peter, M, Wider, G. | | Deposit date: | 2010-04-19 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the conserved ubiquitin-binding motifs (UBMs) of the translesion polymerase iota in complex with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
2KWV
 
 | | Solution Structure of UBM1 of murine Polymerase iota in Complex with Ubiquitin | | Descriptor: | DNA polymerase iota, Ubiquitin | | Authors: | Burschowsky, D, Rudolf, F, Rabut, G, Herrmann, T, Peter, M, Wider, G. | | Deposit date: | 2010-04-20 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the conserved ubiquitin-binding motifs (UBMs) of the translesion polymerase iota in complex with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
3BAZ
 
 | | Structure of hydroxyphenylpyruvate reductase from coleus blumei in complex with NADP+ | | Descriptor: | Hydroxyphenylpyruvate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Janiak, V, Klebe, G, Petersen, M, Heine, A. | | Deposit date: | 2007-11-09 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and substrate docking of a hydroxy(phenyl)pyruvate reductase from the higher plant Coleus blumei Benth
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3BA1
 
 | | Structure of hydroxyphenylpyruvate reductase from coleus blumei | | Descriptor: | Hydroxyphenylpyruvate reductase | | Authors: | Janiak, V, Klebe, G, Petersen, M, Heine, A. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure and substrate docking of a hydroxy(phenyl)pyruvate reductase from the higher plant Coleus blumei Benth.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
184D
 
 | | SELF-ASSOCIATION OF A DNA LOOP CREATES A QUADRUPLEX: CRYSTAL STRUCTURE OF D(GCATGCT) AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3'), MAGNESIUM ION | | Authors: | Leonard, G.A, Zhang, S, Peterson, M.R, Harrop, S.J, Helliwell, J.R, Cruse, W.B.T, Langlois D'Estaintot, B, Kennard, O, Brown, T, Hunter, W.N. | | Deposit date: | 1994-08-10 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Self-association of a DNA loop creates a quadruplex: crystal structure of d(GCATGCT) at 1.8 A resolution.
Structure, 3, 1995
|
|
4EMS
 
 | |
2AX0
 
 | | Hepatitis C Virus NS5b RNA Polymerase in complex with a covalent inhibitor (5x) | | Descriptor: | 5R-(2E-METHYL-3-PHENYL-ALLYL)-3-(BENZENESULFONYLAMINO)-4-OXO-2-THIONOTHIAZOLIDINE, Genome polyprotein, SULFATE ION | | Authors: | Powers, J.P, Piper, D.E, Li, Y, Mayorga, V, Anzola, J, Chen, J.M, Jaen, J.C, Lee, G, Liu, J, Peterson, M.G, Tonn, G.R, Ye, Q, Walker, N.P, Wang, Z. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SAR and Mode of Action of Novel Non-Nucleoside Inhibitors of Hepatitis C NS5b RNA Polymerase.
J.Med.Chem., 49, 2006
|
|
2WCN
 
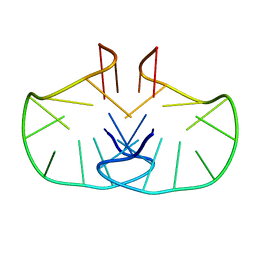 | | Solution structure of an LNA-modified quadruplex | | Descriptor: | DNA (5'-D(*DGP*LCG*DGP*LCG*DTP*DTP*DTP *DTP*DGP*LCG*DGP*LCG)-3') | | Authors: | Nielsen, J.T, Arar, K, Petersen, M. | | Deposit date: | 2009-03-12 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Locked Nucleic Acid Modified Quadruplex: Introducing the V4 Folding Topology.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
1W86
 
 | |
4EVI
 
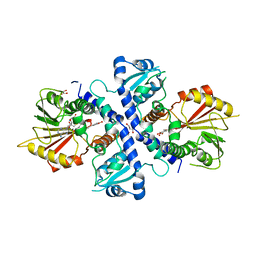 | | Crystal Structure Analysis of Coniferyl Alcohol 9-O-Methyltransferase from Linum Nodiflorum in Complex with Coniferyl Alcohol 9-Methyl Ether and S -Adenosyl-L-Homocysteine | | Descriptor: | 2-methoxy-4-[(1E)-3-methoxyprop-1-en-1-yl]phenol, 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2-methoxyphenol, Coniferyl alcohol 9-O-methyltransferase, ... | | Authors: | Wolters, S, Heine, A, Petersen, M. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Structural analysis of coniferyl alcohol 9-O-methyltransferase from Linum nodiflorum reveals a novel active-site environment.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4E70
 
 | | Crystal Structure Analysis of Coniferyl Alcohol 9-O-Methyltransferase from Linum Nodiflorum in Complex with Coniferyl Alcohol | | Descriptor: | 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2-methoxyphenol, Coniferyl alcohol 9-O-methyltransferase, GLYCEROL | | Authors: | Wolters, S, Heine, A, Petersen, M. | | Deposit date: | 2012-03-16 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6093 Å) | | Cite: | Structural analysis of coniferyl alcohol 9-O-methyltransferase from Linum nodiflorum reveals a novel active-site environment.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2AX1
 
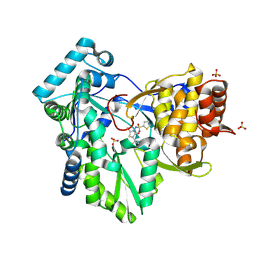 | | Hepatitis C Virus NS5b RNA Polymerase in complex with a covalent inhibitor (5ee) | | Descriptor: | 5R-(3,4-DICHLOROPHENYLMETHYL)-3-(2-THIOPHENESULFONYLAMINO)-4-OXO-2-THIONOTHIAZOLIDINE, Genome polyprotein, SULFATE ION | | Authors: | Powers, J.P, Piper, D.E, Li, Y, Mayorga, V, Anzola, J, Chen, J.M, Jaen, J.C, Lee, G, Liu, J, Peterson, M.G, Tonn, G.R, Ye, Q, Walker, N.P, Wang, Z. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SAR and Mode of Action of Novel Non-Nucleoside Inhibitors of Hepatitis C NS5b RNA Polymerase.
J.Med.Chem., 49, 2006
|
|
2AWZ
 
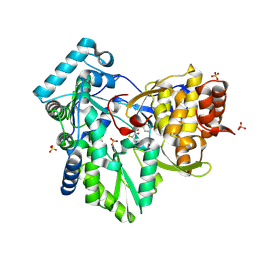 | | Hepatitis C Virus NS5b RNA Polymerase in complex with a covalent inhibitor (5h) | | Descriptor: | 5R-(4-BROMOPHENYLMETHYL)-3-(BENZENESULFONYLAMINO)-4-OXO-2-THIONOTHIAZOLIDINE, Genome polyprotein, SULFATE ION | | Authors: | Powers, J.P, Piper, D.E, Li, Y, Mayorga, V, Anzola, J, Chen, J.M, Jaen, J.C, Lee, G, Liu, J, Peterson, M.G, Tonn, G.R, Ye, Q, Walker, N.P, Wang, Z. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | SAR and Mode of Action of Novel Non-Nucleoside Inhibitors of Hepatitis C NS5b RNA Polymerase.
J.Med.Chem., 49, 2006
|
|
1OCI
 
 | | [3.2.0]bcANA:DNA | | Descriptor: | 5'-D(*CP*TP*GP*A TLBP*AP*TP*GP*CP)-3', 5'-D(*GP*CP*AP*TP*AP*TP*CP*AP*GP)-3' | | Authors: | Tommerholt, H.V, Christensen, N.K, Nielsen, P, Wengel, J, Stein, P.C, Jacobsen, J.P, Petersen, M. | | Deposit date: | 2003-02-07 | | Release date: | 2003-07-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a DsDNA Containing a Bicyclic D-Arabino-Configured Nucleotide Fixed in an O4'-Endo Sugar Conformation
Org.Biomol.Chem., 1, 2003
|
|
