2KA7
 
 | | NMR solution structure of TM0212 at 40 C | | Descriptor: | Glycine cleavage system H protein | | Authors: | Pedrini, B, Herrmann, T, Mohanty, B, Geralt, M, Wilson, I, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-31 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | The J-UNIO protocol for automated protein structure determination by NMR in solution.
J.Biomol.Nmr, 53, 2012
|
|
2JMS
 
 | | NMR Structure of En-6 pheromone from the Antarctic Ciliate Euplotes nobilii | | Descriptor: | Pheromone En-6 | | Authors: | Pedrini, B, Placzek, W.J, Koculi, E, Alimenti, C, LaTerza, A, Luporini, P, Wuthrich, K. | | Deposit date: | 2006-11-29 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Cold-adaptation in Sea-water-borne Signal Proteins: Sequence and NMR Structure of the Pheromone En-6 from the Antarctic Ciliate Euplotes nobilii
J.Mol.Biol., 372, 2007
|
|
2KC6
 
 | | NMR solution structure of the pheromone En-1 of Euplotes nobilii at -1.5 C | | Descriptor: | Mating pheromone En-1 | | Authors: | Pedrini, B, Alimenti, C, Vallesi, A, Luporini, P, Wuthrich, K. | | Deposit date: | 2008-12-17 | | Release date: | 2009-08-04 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Molecular cold-adaptation: Comparative analysis of two homologous families of psychrophilic and mesophilic signal proteins of the protozoan ciliate, Euplotes.
Iubmb Life, 61, 2009
|
|
2KL2
 
 | | NMR solution structure of A2LD1 (gi:13879369) | | Descriptor: | AIG2-like domain-containing protein 1 | | Authors: | Pedrini, B, Serrano, P, Mohanty, B, Geralt, M, Herrmann, T, Wuthrich, K, Wilson, I, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2N2S
 
 | |
2KK2
 
 | | NMR solution structure of the pheromone En-A1 from Euplotes nobilii | | Descriptor: | En-A1 | | Authors: | Pedrini, B, Alimenti, C, Vallesi, A, Luporini, P, Wuthrich, K. | | Deposit date: | 2009-06-15 | | Release date: | 2010-05-12 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Antarctic and Arctic populations of the ciliate Euplotes nobilii show common pheromone-mediated cell-cell signaling and cross-mating.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7AT6
 
 | | Structure of thaumatin collected by femtosecond serial crystallography on a COC membrane | | Descriptor: | L(+)-TARTARIC ACID, R-1,2-PROPANEDIOL, SODIUM ION, ... | | Authors: | Martiel, I, Marsh, M, Vera, L, Huang, C.Y, Olieric, V, Leonarski, P, Nass, K, Padeste, C, Karpik, A, Wang, M, Pedrini, B. | | Deposit date: | 2020-10-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Commissioning results from the SwissMX instrument for fixed target macromolecular crystallography at SwissFEL
To Be Published
|
|
7BET
 
 | | Structure of Ribonucleotide reductase R2 from Escherichia coli collected by femtosecond serial crystallography on a COC membrane | | Descriptor: | FE (III) ION, Ribonucleoside-diphosphate reductase 1 subunit beta | | Authors: | Aurelius, O, John, J, Martiel, I, Marsh, M, Vera, L, Huang, C.Y, Olieric, V, Leonarski, P, Nass, K, Padeste, C, Karpik, A, Hogbom, M, Wang, M, Pedrini, B. | | Deposit date: | 2020-12-24 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Commissioning results from the SwissMX instrument for fixed target macromolecular crystallography at SwissFEL
To Be Published
|
|
2RNK
 
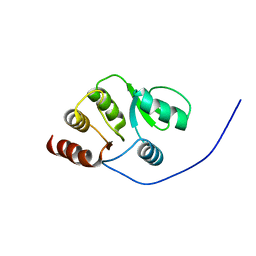 | | NMR structure of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B.W, Wilson, I.A, Stevens, R.C, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-11 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2JZD
 
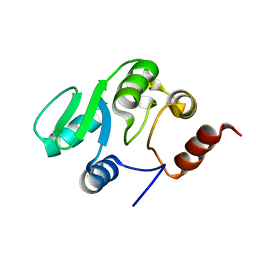 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2JZE
 
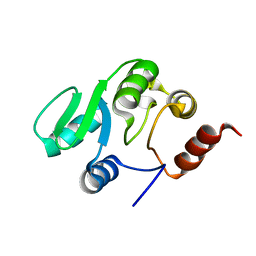 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3, single conformer closest to the mean coordinates of an ensemble of twenty energy minimized conformers | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2JZF
 
 | | NMR Conformer closest to the mean coordinates of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2KA0
 
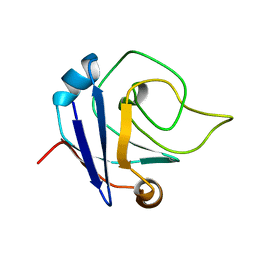 | | NMR structure of the protein TM1367 | | Descriptor: | uncharacterized protein TM1367 | | Authors: | Mohanty, B, Pedrini, B, Serrano, P, Geralt, M, Horst, R, Herrmann, T, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-27 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures for the proteins TM1112 and TM1367.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2KA5
 
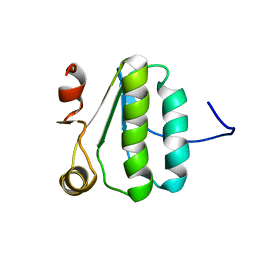 | | NMR Structure of the protein TM1081 | | Descriptor: | Putative anti-sigma factor antagonist TM_1081 | | Authors: | Serrano, P, Geralt, M, Mohanty, B, Pedrini, B, Horst, R, Wuthrich, K, Wilson, I, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2K9Z
 
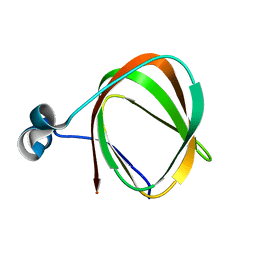 | | NMR structure of the protein TM1112 | | Descriptor: | uncharacterized protein TM1112 | | Authors: | Mohanty, B, Pedrini, B, Serrano, P, Geralt, M, Horst, R, Herrmann, T, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-28 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures for the proteins TM1112 and TM1367.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2MU2
 
 | | NMR structure of the cap domain of NP_346487.1, a putative phosphoglycolate phosphatase from Streptococcus pneumoniae TIGR4 | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Jaudzems, K, Serrano, P, Pedrini, B, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-09-03 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | J-UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4.
J.Biomol.Nmr, 61, 2015
|
|
2MSN
 
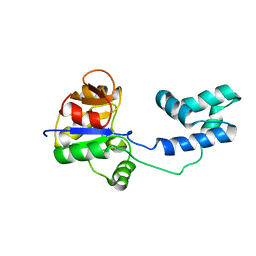 | | NMR structure of a putative phosphoglycolate phosphatase (NP_346487.1) from Streptococcus pneumoniae TIGR4 | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Jaudzems, K, Serrano, P, Pedrini, B, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | J-UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4.
J.Biomol.Nmr, 61, 2015
|
|
2MU1
 
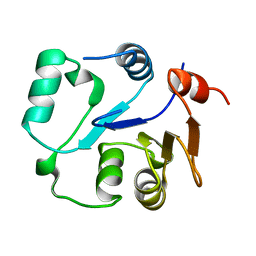 | | NMR structure of the core domain of NP_346487.1, a putative phosphoglycolate phosphatase from Streptococcus pneumoniae TIGR4 | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Jaudzems, K, Serrano, P, Pedrini, B, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-09-03 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | J-UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4.
J.Biomol.Nmr, 61, 2015
|
|
2KAF
 
 | | Solution structure of the SARS-unique domain-C from the nonstructural protein 3 (nsp3) of the severe acute respiratory syndrome coronavirus | | Descriptor: | Non-structural protein 3 | | Authors: | Johnson, M.A, Mohanty, B, Pedrini, B, Serrano, P, Chatterjee, A, Herrmann, T, Joseph, J, Saikatendu, K, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-11-05 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding.
J.Mol.Biol., 400, 2010
|
|
2LYY
 
 | | NMR structure of the protein NB7890A from Shewanella sp | | Descriptor: | Uncharacterized protein | | Authors: | Serrano, P, Geralt, M, Pedrini, B, Wuthrich, K, Horst, R, Augustyniak, W, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein NB7890A from Shewanella sp
To be Published
|
|
