4ZCE
 
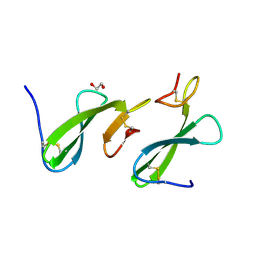 | | Crystal Structure of the dust mite allergen Der p 23 from Dermatophagoides pteronyssinus | | Descriptor: | 1,2-ETHANEDIOL, Dust mite allergen | | Authors: | Pedersen, L.C, Mueller, G.A, Randall, T.A, Glesner, J, Perera, L, Edwards, L.L, Chapman, M.D, London, R.E, Pomes, A. | | Deposit date: | 2015-04-15 | | Release date: | 2015-11-25 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Serological, genomic and structural analyses of the major mite allergen Der p 23.
Clin Exp Allergy, 46, 2016
|
|
8VH7
 
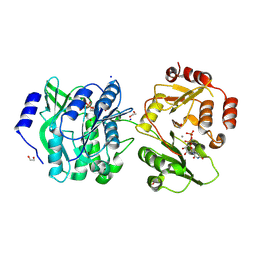 | | Crystal structure of heparosan synthase 2 from Pasteurella multocida at 1.98 A | | Descriptor: | 1,2-ETHANEDIOL, Heparosan synthase B, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Liu, J, Stancanelli, E, Krahn, J.M. | | Deposit date: | 2023-12-31 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Structural and Functional Analysis of Heparosan Synthase 2 from Pasteurella multocida to Improve the Synthesis of Heparin
Acs Catalysis, 14, 2024
|
|
4Q5R
 
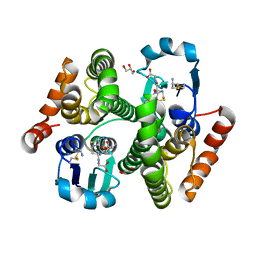 | |
4Q5Q
 
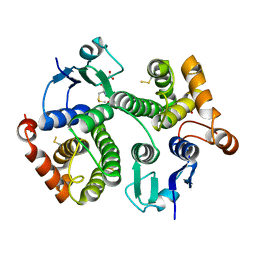 | |
4Q5N
 
 | |
4Q5F
 
 | |
4QGO
 
 | | Crystal structure of NucA from Streptococcus agalactiae with no metal bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-entry nuclease (Competence-specific nuclease), ... | | Authors: | Pedersen, L.C, Moon, A.F, Gaudu, P. | | Deposit date: | 2014-05-23 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of the virulence factor nuclease A from Streptococcus agalactiae.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1KNY
 
 | | KANAMYCIN NUCLEOTIDYLTRANSFERASE | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, KANAMYCIN A, KANAMYCIN NUCLEOTIDYLTRANSFERASE, ... | | Authors: | Pedersen, L.C, Benning, M.M, Holden, H.M. | | Deposit date: | 1995-07-07 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural investigation of the antibiotic and ATP-binding sites in kanamycin nucleotidyltransferase.
Biochemistry, 34, 1995
|
|
1EFH
 
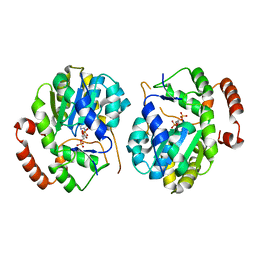 | |
1FGG
 
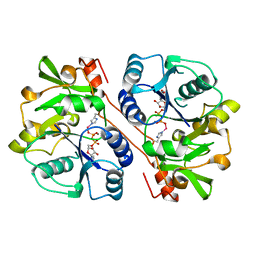 | | CRYSTAL STRUCTURE OF 1,3-GLUCURONYLTRANSFERASE I (GLCAT-I) COMPLEXED WITH GAL-GAL-XYL, UDP, AND MN2+ | | Descriptor: | GLUCURONYLTRANSFERASE I, MANGANESE (II) ION, UNKNOWN ATOM OR ION, ... | | Authors: | Pedersen, L.C, Tsuchida, K, Kitagawa, H, Sugahara, K, Darden, T.A. | | Deposit date: | 2000-07-28 | | Release date: | 2001-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Heparan/chondroitin sulfate biosynthesis. Structure and mechanism of human glucuronyltransferase I.
J.Biol.Chem., 275, 2000
|
|
8SLR
 
 | | Crystal Structure of mouse TRAIL | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pedersen, L.C, Xu, D. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Heparan sulfate promotes TRAIL-induced tumor cell apoptosis.
Elife, 12, 2024
|
|
1RZT
 
 | | Crystal structure of DNA polymerase lambda complexed with a two nucleotide gap DNA molecule | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*GP*GP*CP*AP*AP*CP*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*G)-3', ... | | Authors: | Pedersen, L.C, Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Blanco, L, Kunkel, T.A. | | Deposit date: | 2003-12-29 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural solution for the DNA polymerase lambda-dependent repair of DNA gaps with minimal homology.
Mol.Cell, 13, 2004
|
|
1QXM
 
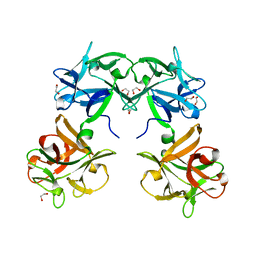 | | Crystal structure of a hemagglutinin component (HA1) from type C Clostridium botulinum | | Descriptor: | 1,2-ETHANEDIOL, HA1 | | Authors: | Inoue, K, Sobhany, M, Transue, T.R, Oguma, K, Pedersen, L.C, Negishi, M. | | Deposit date: | 2003-09-08 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis by X-ray crystallography and calorimetry of a haemagglutinin component (HA1) of the progenitor toxin from Clostridium botulinum.
Microbiology, 149, 2003
|
|
3C2M
 
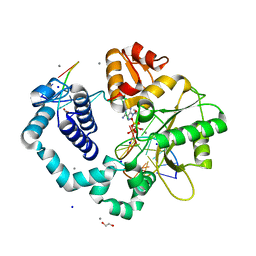 | | Ternary complex of DNA POLYMERASE BETA with a G:dAPCPP mismatch in the active site | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]adenosine, DNA (5'-D(*DCP*DCP*DGP*DAP*DCP*DGP*DGP*DCP*DGP*DCP*DAP*DTP*DCP*DAP*DGP*DC)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of DNA polymerase beta with active-site mismatches suggest a transient abasic site intermediate during misincorporation.
Mol.Cell, 30, 2008
|
|
3C2K
 
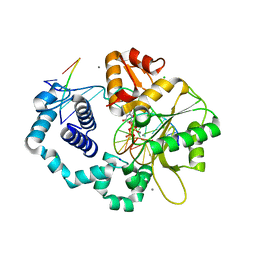 | | DNA POLYMERASE BETA with a gapped DNA substrate and DUMPNPP with Manganese in the active site | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*DCP*DCP*DGP*DAP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DCP*DAP*DGP*DC)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of DNA polymerase beta with active-site mismatches suggest a transient abasic site intermediate during misincorporation.
Mol.Cell, 30, 2008
|
|
3C5F
 
 | | Structure of a binary complex of the R517A Pol lambda mutant | | Descriptor: | DNA (5'-D(*DCP*DAP*DGP*DTP*DAP*DC)-3'), DNA (5'-D(*DCP*DGP*DGP*DCP*DCP*DGP*DTP*DAP*DCP*DTP*DG)-3'), DNA (5'-D(P*DGP*DCP*DCP*DG)-3'), ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Foley, M.C, Pedersen, L.C, Schlick, T, Kunkel, T.A. | | Deposit date: | 2008-01-31 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate-induced DNA strand misalignment during catalytic cycling by DNA polymerase lambda.
Embo Rep., 9, 2008
|
|
3BD9
 
 | | human 3-O-sulfotransferase isoform 5 with bound PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Xu, D, Moon, A.F, Song, D, Liu, J, Pedersen, L.C. | | Deposit date: | 2007-11-14 | | Release date: | 2008-01-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering sulfotransferases to modify heparan sulfate.
Nat.Chem.Biol., 4, 2008
|
|
8DB4
 
 | |
6BKG
 
 | | Human LigIV catalytic domain with bound DNA-adenylate intermediate in closed conformation | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Moon, A.F, Tumbale, P.P, Schellenberg, M.J, Williams, R.S, Williams, J.G, Kunkel, T.A, Pedersen, L.C, Bebenek, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structures of DNA-bound human ligase IV catalytic core reveal insights into substrate binding and catalysis.
Nat Commun, 9, 2018
|
|
6BKF
 
 | | Lysyl-adenylate form of human LigIV catalytic domain with bound DNA substrate in open conformation | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*C)-3'), DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Moon, A.F, Tumbale, P.P, Schellenberg, M.J, Williams, R.S, Williams, J.G, Kunkel, T.A, Pedersen, L.C, Bebenek, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structures of DNA-bound human ligase IV catalytic core reveal insights into substrate binding and catalysis.
Nat Commun, 9, 2018
|
|
6TYW
 
 | |
6TYZ
 
 | |
6TYV
 
 | |
6TYX
 
 | |
6TYU
 
 | |
