7N8U
 
 | |
4WR3
 
 | | Y274F alanine racemase from E. coli | | Descriptor: | Alanine racemase, biosynthetic, GLYCEROL, ... | | Authors: | Squire, C.J, Yosaatmadja, Y, Patrick, W.M. | | Deposit date: | 2014-10-23 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic and Evolutionary Insights from the Reciprocal Promiscuity of Two Pyridoxal Phosphate-dependent Enzymes.
J.Biol.Chem., 291, 2016
|
|
4XBJ
 
 | | Y274F alanine racemase from E. coli inhibited by l-ala-p | | Descriptor: | Alanine racemase, biosynthetic, SULFATE ION, ... | | Authors: | Squire, C.J, Yosaatmadja, Y, Patrick, W.M. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic and Evolutionary Insights from the Reciprocal Promiscuity of Two Pyridoxal Phosphate-dependent Enzymes.
J.Biol.Chem., 291, 2016
|
|
7KB0
 
 | | O-acety-L-homoserine aminocarboxypropyltransferase (MetY) from Thermotoga maritima with pyridoxal-5-phosphate (PLP) bound in the internal aldimine state | | Descriptor: | O-acetyl-L-homoserine sulfhydrylase | | Authors: | Brewster, J.L, Pachl, P, Squire, C, Selmer, M, Patrick, W.M. | | Deposit date: | 2020-10-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures and kinetics of Thermotoga maritima MetY reveal new insights into the predominant sulfurylation enzyme of bacterial methionine biosynthesis.
J.Biol.Chem., 296, 2021
|
|
7KB1
 
 | | Complex of O-acety-L-homoserine aminocarboxypropyltransferase (MetY) from Thermotoga maritima and a key reaction intermediate | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, MAGNESIUM ION, O-acetyl-L-homoserine sulfhydrylase, ... | | Authors: | Brewster, J.L, Pachl, P, Squire, C, Selmer, M, Patrick, W.M. | | Deposit date: | 2020-10-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures and kinetics of Thermotoga maritima MetY reveal new insights into the predominant sulfurylation enzyme of bacterial methionine biosynthesis.
J.Biol.Chem., 296, 2021
|
|
7R7M
 
 | |
7R9B
 
 | |
7RCQ
 
 | |
6BI6
 
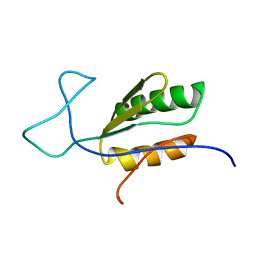 | | Solution NMR structure of uncharacterized protein YejG | | Descriptor: | Uncharacterized protein YejG | | Authors: | Mohanty, B, Finn, T.J, Macindoe, I, Zhong, J, Patrick, W.M, Mackay, J.P. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The uncharacterized bacterial protein YejG has the same architecture as domain III of elongation factor G.
Proteins, 87, 2019
|
|
2KZH
 
 | |
7RGC
 
 | |
7RMN
 
 | |
7RPN
 
 | |
2PC8
 
 | | E292Q mutant of EXO-B-(1,3)-Glucanase from Candida Albicans in complex with two separately bound glucopyranoside units at 1.8 A | | Descriptor: | Hypothetical protein XOG1, beta-D-glucopyranose | | Authors: | Cutfield, S.M, Cutfield, J.F, Patrick, W.M. | | Deposit date: | 2007-03-29 | | Release date: | 2008-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Carbohydrate binding sites in Candida albicans exo-beta-1,3-glucanase and the role of the Phe-Phe 'clamp' at the active site entrance.
Febs J., 277, 2010
|
|
2PF0
 
 | |
2PBO
 
 | |
4YI7
 
 | |
5A5W
 
 | | Crystal structure of Salmonella enterica HisA D7N D176A with ProFAR | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, [(2R,3S,4R,5R)-5-[4-aminocarbonyl-5-[(E)-[[(2R,3R,4S,5R)-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]amino]methylideneamino]imidazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Soderholm, A, Guo, X, Newton, M.S, Evans, G.B, Nasvall, J, Patrick, W.M, Selmer, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two-Step Ligand Binding in a Beta/Alpha8 Barrel Enzyme -Substrate-Bound Structures Shed New Light on the Catalytic Cycle of Hisa
J.Biol.Chem., 290, 2015
|
|
5AHI
 
 | | Crystal structure of salmonalla enterica HisA mutant D7N with ProFAR | | Descriptor: | 1-(5-PHOSPHORIBOSYL)-5-[(5-PHOSPHORIBOSYLAMINO) METHYLIDENE AMINO] IMIDAZOLE-4-CARBOXAMIDE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Soderholm, A, Guo, X, Newton, M.S, Evans, G.B, Nasvall, J, Patrick, W.M, Selmer, M. | | Deposit date: | 2015-02-06 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structure and Mechanism of Hisa from Salmonella Enterica
To be Published
|
|
5AHF
 
 | | Crystal structure of Salmonella enterica HisA D7N with ProFAR | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, GLYCEROL, [(2R,3S,4R,5R)-5-[4-aminocarbonyl-5-[(E)-[[(2R,3R,4S,5R)-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]amino]methylideneamino]imidazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Soderholm, A, Guo, X, Newton, M.S, Evans, G.B, Nasvall, J, Patrick, W.M, Selmer, M. | | Deposit date: | 2015-02-05 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Two-Step Ligand Binding in a Beta/Alpha8 Barrel Enzyme -Substrate-Bound Structures Shed New Light on the Catalytic Cycle of Hisa
J.Biol.Chem., 290, 2015
|
|
5AHE
 
 | | Crystal structure of Salmonella enterica HisA | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Soderholm, A, Guo, X, Newton, M.S, Evans, G.B, Nasvall, J, Patrick, W.M, Selmer, M. | | Deposit date: | 2015-02-05 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two-Step Ligand Binding in a Beta/Alpha8 Barrel Enzyme -Substrate-Bound Structures Shed New Light on the Catalytic Cycle of Hisa
J.Biol.Chem., 290, 2015
|
|
4ITG
 
 | | P113S mutant of E. coli Cystathionine beta-lyase MetC | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cystathionine beta-lyase MetC | | Authors: | Squire, C.J, Yosaatmadja, Y, Soo, V.W.C, Patrick, W.M. | | Deposit date: | 2013-01-18 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mechanistic and Evolutionary Insights from the Reciprocal Promiscuity of Two Pyridoxal Phosphate-dependent Enzymes.
J.Biol.Chem., 291, 2016
|
|
4ITX
 
 | | P113S mutant of E. coli Cystathionine beta-lyase MetC inhibited by reaction with L-Ala-P | | Descriptor: | CALCIUM ION, Cystathionine beta-lyase MetC, {1-[(3-HYDROXY-METHYL-5-PHOSPHONOOXY-METHYL-PYRIDIN-4-YLMETHYL)-AMINO]-ETHYL}-PHOSPHONIC ACID | | Authors: | Squire, C.J, Yosaatmadja, Y, Soo, V.W.C, Patrick, W.M. | | Deposit date: | 2013-01-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Mechanistic and Evolutionary Insights from the Reciprocal Promiscuity of Two Pyridoxal Phosphate-dependent Enzymes.
J.Biol.Chem., 291, 2016
|
|
4GTN
 
 | |
