6W4Z
 
 | | Galectin-8N terminal domain in complex with Methyl 3-O-[3-O-benzyloxy]-malonyl-beta-D-galactopyranoside | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Galectin-8, ... | | Authors: | Patel, B, Kishor, C, Blanchard, H. | | Deposit date: | 2020-03-12 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Rational Design and Synthesis of Methyl-beta-d-galactomalonyl Phenyl Esters as Potent Galectin-8 N Antagonists.
J.Med.Chem., 63, 2020
|
|
3HJ6
 
 | | Structure of Halothermothrix orenii fructokinase (FRK) | | Descriptor: | Fructokinase | | Authors: | Chua, T.K, Seetharaman, J, Kasprzak, J.M, Ng, C, Patel, B.K, Love, C, Bujnicki, J.M, Sivaraman, J. | | Deposit date: | 2009-05-21 | | Release date: | 2010-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a fructokinase homolog from Halothermothrix orenii
J.Struct.Biol., 171, 2010
|
|
2B0H
 
 | | Solution structure of VBS3 fragment of talin | | Descriptor: | Talin-1 | | Authors: | Gingras, A.R, Vogel, K.P, Steinhoff, H.J, Ziegler, W.H, Patel, B, Emsley, J, Critchley, D.R, Roberts, G.C, Barsukov, I.L. | | Deposit date: | 2005-09-14 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic Characterization of a Vinculin Binding Site in the Talin Rod
Biochemistry, 45, 2006
|
|
1U89
 
 | | Solution structure of VBS2 fragment of talin | | Descriptor: | Talin 1 | | Authors: | Fillingham, I, Gingras, A.R, Papagrigoriou, E, Patel, B, Emsley, J, Roberts, G.C.K, Critchley, D.R, Barsukov, I.L. | | Deposit date: | 2004-08-05 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head.
Structure, 13, 2005
|
|
1U6H
 
 | | Vinculin head (0-258) in complex with the talin vinculin binding site 2 (849-879) | | Descriptor: | Talin, Vinculin | | Authors: | Fillingham, I, Gingras, A.R, Papagrigoriou, E, Patel, B, Emsley, J, Roberts, G.C.K, Critchley, D.R, Barsukov, I.L. | | Deposit date: | 2004-07-30 | | Release date: | 2005-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head.
Structure, 13, 2005
|
|
1ZX9
 
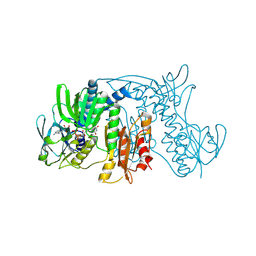 | | Crystal Structure of Tn501 MerA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Mercuric reductase | | Authors: | Dong, A, Ledwidge, R, Patel, B, Fiedler, D, Falkowski, M, Zelikova, J, Summers, A.O, Pai, E.F, Miller, S.M. | | Deposit date: | 2005-06-07 | | Release date: | 2005-07-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NmerA, the Metal Binding Domain of Mercuric Ion Reductase, Removes Hg(2+) from Proteins, Delivers It to the Catalytic Core, and Protects Cells under Glutathione-Depleted Conditions
Biochemistry, 44, 2005
|
|
1ZK7
 
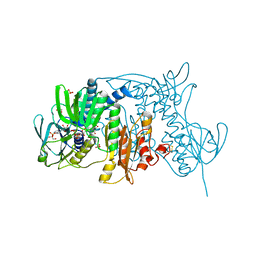 | | Crystal Structure of Tn501 MerA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Mercuric reductase, ... | | Authors: | Dong, A, Ledwidge, R, Patel, B, Fiedler, D, Falkowski, M, Zelikova, J, Summers, A.O, Pai, E.F, Miller, S.M. | | Deposit date: | 2005-05-02 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | NmerA, the Metal Binding Domain of Mercuric Ion Reductase, Removes Hg(2+) from Proteins, Delivers It to the Catalytic Core, and Protects Cells under Glutathione-Depleted Conditions
Biochemistry, 44, 2005
|
|
4QQS
 
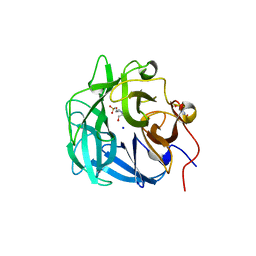 | | Crystal structure of a thermostable family-43 glycoside hydrolase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Glycoside hydrolase family 43, SODIUM ION | | Authors: | Hassan, N, Kori, L.D, Patel, B.K.C, Divne, C, Tan, T.C. | | Deposit date: | 2014-06-29 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution crystal structure of a polyextreme GH43 glycosidase from Halothermothrix orenii with alpha-L-arabinofuranosidase activity.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
3TA9
 
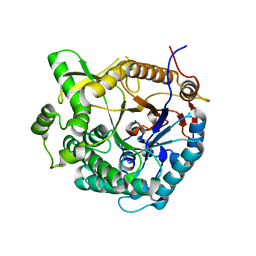 | | beta-Glucosidase A from the halothermophile H. orenii | | Descriptor: | Glycoside hydrolase family 1, NICKEL (II) ION | | Authors: | Hofmann, A, Wang, C.K, Kori, L.D, Patel, B.K. | | Deposit date: | 2011-08-03 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Expression, purification and preliminary crystallographic analysis of the recombinant (beta)-glucosidase (BglA) from the halothermophile Halothermothrix orenii
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3BCF
 
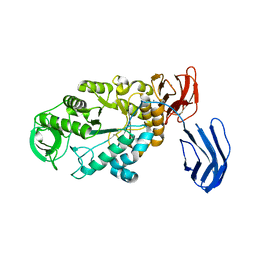 | | Alpha-amylase B from Halothermothrix orenii | | Descriptor: | Alpha amylase, catalytic region, CALCIUM ION, ... | | Authors: | Tan, T.-C, Mijts, B.N, Swaminathan, K, Patel, B.K.C, Divne, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Polyextremophilic alpha-Amylase AmyB from Halothermothrix orenii: Details of a Productive Enzyme-Substrate Complex and an N Domain with a Role in Binding Raw Starch
J.Mol.Biol., 378, 2008
|
|
4PTX
 
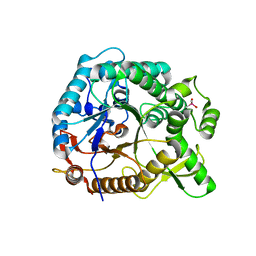 | | Halothermothrix orenii beta-glucosidase A, glucose complex | | Descriptor: | CACODYLATE ION, Glycoside hydrolase family 1, beta-D-glucopyranose | | Authors: | Hassan, N, Nguyen, T.H, Kori, L.D, Patel, B.K.C, Haltrich, D, Divne, C, Tan, T.C. | | Deposit date: | 2014-03-11 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and structural characterization of a thermostable beta-glucosidase from Halothermothrix orenii for galacto-oligosaccharide synthesis.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4PTV
 
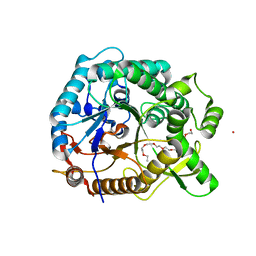 | | Halothermothrix orenii beta-glucosidase A, thiocellobiose complex | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CESIUM ION, Glycoside hydrolase family 1, ... | | Authors: | Hassan, N, Nguyen, T.H, Kori, L.D, Patel, B.K.C, Haltrich, D, Divne, C, Tan, T.C. | | Deposit date: | 2014-03-11 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural characterization of a thermostable beta-glucosidase from Halothermothrix orenii for galacto-oligosaccharide synthesis.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3BCD
 
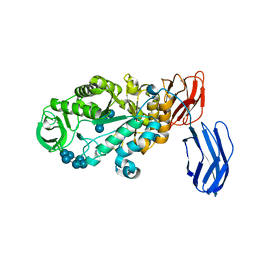 | | Alpha-amylase B in complex with maltotetraose and alpha-cyclodextrin | | Descriptor: | Alpha amylase, catalytic region, CALCIUM ION, ... | | Authors: | Tan, T.-C, Mijts, B.N, Swaminathan, K, Patel, B.K.C, Divne, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Polyextremophilic alpha-Amylase AmyB from Halothermothrix orenii: Details of a Productive Enzyme-Substrate Complex and an N Domain with a Role in Binding Raw Starch
J.Mol.Biol., 378, 2008
|
|
4PTW
 
 | | Halothermothrix orenii beta-glucosidase A, 2-deoxy-2-fluoro-glucose complex | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-glucopyranose, Glycoside hydrolase family 1, TETRAETHYLENE GLYCOL | | Authors: | Hassan, N, Nguyen, T.H, Kori, L.D, Patel, B.K.C, Haltrich, D, Divne, C, Tan, T.C. | | Deposit date: | 2014-03-11 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural characterization of a thermostable beta-glucosidase from Halothermothrix orenii for galacto-oligosaccharide synthesis.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3BC9
 
 | | Alpha-amylase B in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Tan, T.-C, Mijts, B.N, Swaminathan, K, Patel, B.K.C, Divne, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of the Polyextremophilic alpha-Amylase AmyB from Halothermothrix orenii: Details of a Productive Enzyme-Substrate Complex and an N Domain with a Role in Binding Raw Starch
J.Mol.Biol., 378, 2008
|
|
4CFE
 
 | | Structure of full length human AMPK in complex with a small molecule activator, a benzimidazole derivative (991) | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Carmena, D, Bright, N.J, Haire, L.F, Underwood, E, Patel, B.R, Heath, R.B, Walker, P.A, Hallen, S, Giordanetto, F, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.023 Å) | | Cite: | Structural Basis of Ampk Regulation by Small Molecule Activators.
Nat.Commun., 4, 2013
|
|
4CFF
 
 | | Structure of full length human AMPK in complex with a small molecule activator, a thienopyridone derivative (A-769662) | | Descriptor: | 3-[4-(2-hydroxyphenyl)phenyl]-4-oxidanyl-6-oxidanylidene-7H-thieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Carmena, D, Bright, N.J, Haire, L.F, Underwood, E, Patel, B.R, Heath, R.B, Walker, P.A, Hallen, S, Giordanetto, F, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.924 Å) | | Cite: | Structural Basis of Ampk Regulation by Small Molecule Activators.
Nat.Commun., 4, 2013
|
|
