4O8F
 
 | | Crystal Structure of the complex between PPARgamma mutant R357A and rosiglitazone | | Descriptor: | 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Chiaraluce, R, Consalvi, V, Lori, C, Loiodice, F, Laghezza, A, Pasquo, A, Cervoni, L, Aschi, M. | | Deposit date: | 2013-12-27 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the transactivation deficiency of the human PPAR gamma F360L mutant associated with familial partial lipodystrophy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2VNE
 
 | | The X-ray structure of Norcoclaurine synthase from Thalictrum flavum | | Descriptor: | S-NORCOCLAURINE SYNTHASE | | Authors: | Ilari, A, Franceschini, S, Boffi, A, Bonamore, A, Pasquo, A. | | Deposit date: | 2008-02-04 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis of Enzymatic S-Norcoclaurine Biosynthesis.
J.Biol.Chem., 284, 2009
|
|
1GTM
 
 | | STRUCTURE OF GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE, SULFATE ION | | Authors: | Yip, K.S.P, Stillman, T.J, Britton, K.L, Pasquo, A, Rice, D.W. | | Deposit date: | 1996-08-22 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of Pyrococcus furiosus glutamate dehydrogenase reveals a key role for ion-pair networks in maintaining enzyme stability at extreme temperatures.
Structure, 3, 1995
|
|
5HEL
 
 | | Crystal structure of the N-terminus Y153H bromodomain mutant of human BRD2 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2 | | Authors: | Tallant, C, Lori, C, Pasquo, A, Chiaraluce, R, Consalvi, V, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2016-01-06 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the N-terminus Y153H bromodomain mutant of human BRD2
To Be Published
|
|
4L96
 
 | | Structure of the complex between the F360L PPARgamma mutant and the ligand LT175 (space group I222) | | Descriptor: | (2S)-2-(biphenyl-4-yloxy)-3-phenylpropanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Chiaraluce, R, Consalvi, V, Pasquo, A, Lori, C, Laghezza, A, Loiodice, F. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis of the transactivation deficiency of the human PPAR gamma F360L mutant associated with familial partial lipodystrophy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4L98
 
 | | Crystal structure of the complex of F360L PPARgamma mutant with the ligand LT175 | | Descriptor: | (2S)-2-(biphenyl-4-yloxy)-3-phenylpropanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Consalvi, V, Chiaraluce, R, Pasquo, A, Capelli, D, Loiodice, F, Laghezza, A, Lori, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis of the transactivation deficiency of the human PPAR gamma F360L mutant associated with familial partial lipodystrophy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1Q8R
 
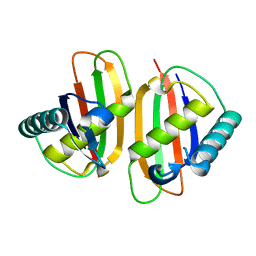 | | Structure of E.coli RusA Holliday junction resolvase | | Descriptor: | Crossover junction endodeoxyribonuclease rusA | | Authors: | Rafferty, J.B, Bolt, E.L, Muranova, T.A, Sedelnikova, S.E, Leonard, P, Pasquo, A, Baker, P.J, Rice, D.W, Sharples, G.J, Lloyd, R.G. | | Deposit date: | 2003-08-22 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | The structure of Escherichia coli RusA endonuclease reveals a new Holliday junction DNA binding fold
Structure, 11, 2003
|
|
5HEM
 
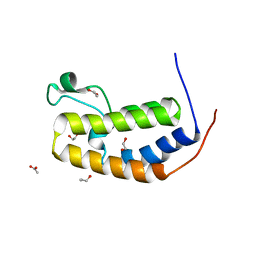 | | Crystal structure of the N-terminus D161Y bromodomain mutant of human BRD2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Bromodomain-containing protein 2, ... | | Authors: | Tallant, C, Lori, C, Pasquo, A, Chiaraluce, R, Consalvi, V, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2016-01-06 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the N-terminus D161Y bromodomain mutant of human BRD2
To Be Published
|
|
5HFQ
 
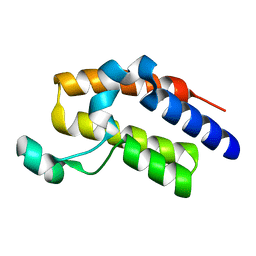 | | Crystal structure of the second bromodomain Q443H mutant of human BRD2 | | Descriptor: | Bromodomain-containing protein 2 | | Authors: | Tallant, C, Lori, C, Pasquo, A, Chiaraluce, R, Consalvi, V, Fonseca, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2016-01-07 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the second bromodomain Q443H mutant of human BRD2
To Be Published
|
|
5HFR
 
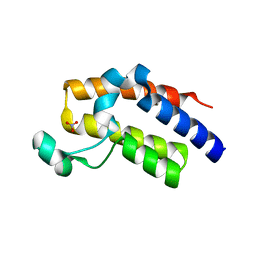 | | Crystal structure of the second bromodomain H395R mutant of human BRD3 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, NITRATE ION | | Authors: | Tallant, C, Lori, C, Pasquo, A, Chiaraluce, R, Consalvi, V, Fonseca, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2016-01-07 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the second bromodomain H395R mutant of human BRD3
To Be Published
|
|
5HEN
 
 | | Crystal structure of the N-terminus R100L bromodomain mutant of human BRD2 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2 | | Authors: | Tallant, C, Lori, C, Pasquo, A, Chiaraluce, R, Consalvi, V, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2016-01-06 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the N-terminus R100L bromodomain mutant of human BRD2
To Be Published
|
|
