5GOX
 
 | | Eukaryotic Rad50 Functions as A Rod-shaped Dimer | | 分子名称: | DNA repair protein RAD50, GLYCEROL, ZINC ION | | 著者 | Park, Y.B, Hohl, M, Padjasek, M, Jeong, E, Jin, K.S, Krezel, A, Petrini, J.H.J, Cho, Y. | | 登録日 | 2016-07-30 | | 公開日 | 2017-02-01 | | 最終更新日 | 2017-03-15 | | 実験手法 | X-RAY DIFFRACTION (2.405 Å) | | 主引用文献 | Eukaryotic Rad50 functions as a rod-shaped dimer
Nat. Struct. Mol. Biol., 24, 2017
|
|
1F3V
 
 | | Crystal structure of the complex between the N-terminal domain of TRADD and the TRAF domain of TRAF2 | | 分子名称: | TUMOR NECROSIS FACTOR RECEPTOR TYPE 1 ASSOCIATED DEATH DOMAIN PROTEIN, TUMOR NECROSIS FACTOR RECEPTOR-ASSOCIATED PROTEIN | | 著者 | Park, Y.C, Ye, H, Hsia, C, Segal, D, Rich, R, Liou, H.-C, Myszka, D, Wu, H. | | 登録日 | 2000-06-06 | | 公開日 | 2000-09-06 | | 最終更新日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A novel mechanism of TRAF signaling revealed by structural and functional analyses of the TRADD-TRAF2 interaction.
Cell(Cambridge,Mass.), 101, 2000
|
|
2Z2R
 
 | | Nucleosome assembly proteins I (NAP-1, 74-365) | | 分子名称: | Nucleosome assembly protein | | 著者 | Park, Y.J, Luger, K. | | 登録日 | 2007-05-25 | | 公開日 | 2008-03-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | A beta-hairpin comprising the nuclear localization sequence sustains the self-associated states of nucleosome assembly protein 1
J.Mol.Biol., 375, 2008
|
|
5ZZ6
 
 | | Redox-sensing transcriptional repressor Rex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Redox-sensing transcriptional repressor Rex 1 | | 著者 | Park, Y.W, Jang, Y.Y, Joo, H.K, Lee, J.Y. | | 登録日 | 2018-05-30 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Analysis of Redox-sensing Transcriptional Repressor Rex from Thermotoga maritima
Sci Rep, 8, 2018
|
|
5ZZ7
 
 | | Redox-sensing transcriptional repressor Rex | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Redox-sensing transcriptional repressor Rex 1 | | 著者 | Park, Y.W, Jang, Y.Y, Joo, H.K, Lee, J.Y. | | 登録日 | 2018-05-30 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural Analysis of Redox-sensing Transcriptional Repressor Rex from Thermotoga maritima
Sci Rep, 8, 2018
|
|
5ZZ5
 
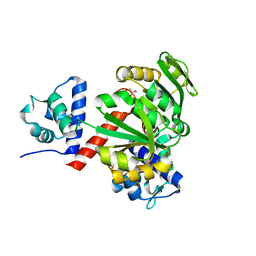 | | Redox-sensing transcriptional repressor Rex | | 分子名称: | GLYCEROL, Redox-sensing transcriptional repressor Rex | | 著者 | Park, Y.W, Jang, Y.Y, Joo, H.K, Lee, J.Y. | | 登録日 | 2018-05-30 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Analysis of Redox-sensing Transcriptional Repressor Rex from Thermotoga maritima
Sci Rep, 8, 2018
|
|
7JV4
 
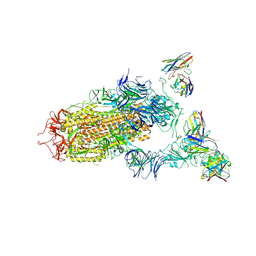 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody (one RBD open) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2H13 Fab heavy chain, ... | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-20 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-01-04 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JZL
 
 | |
7JZN
 
 | |
7JV6
 
 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody (closed conformation) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2H13 Fab heavy chain, ... | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-20 | | 公開日 | 2020-10-14 | | 最終更新日 | 2021-06-23 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JVC
 
 | | SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2A4 Fab heavy chain, ... | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-20 | | 公開日 | 2020-10-14 | | 最終更新日 | 2021-06-23 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
2N40
 
 | |
7JXC
 
 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | 分子名称: | NONAETHYLENE GLYCOL, S2H14 antigen-binding (Fab) fragment | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-27 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JXD
 
 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | 分子名称: | S2A4 antigen-binding (Fab) fragment | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-27 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
2AYU
 
 | |
7JV2
 
 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody Fab fragment (local refinement of the receptor-binding motif and Fab variable domains) | | 分子名称: | S2H13 Fab heavy chain, S2H13 Fab light chain, Spike glycoprotein | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-20 | | 公開日 | 2020-10-14 | | 最終更新日 | 2021-01-27 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JVA
 
 | | SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment (local refinement of the receptor-binding domain and Fab variable domains) | | 分子名称: | S2A4 Fab heavy chain, S2A4 Fab light chain, Spike glycoprotein, ... | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-20 | | 公開日 | 2020-10-14 | | 最終更新日 | 2021-01-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JZM
 
 | |
7JZU
 
 | |
4OK7
 
 | | Structure of bacteriophage SPN1S endolysin from Salmonella typhimurium | | 分子名称: | Endolysin, GLYCEROL, SULFATE ION | | 著者 | Park, Y, Lim, J, Kong, M, Ryu, S, Rhee, S. | | 登録日 | 2014-01-22 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of bacteriophage SPN1S endolysin reveals an unusual two-module fold for the peptidoglycan lytic and binding activity.
Mol.Microbiol., 92, 2014
|
|
4RSX
 
 | |
4RSW
 
 | |
6N38
 
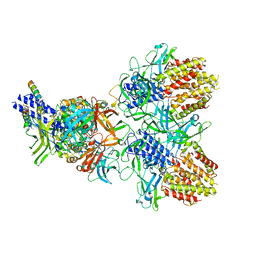 | | Structure of the type VI secretion system TssK-TssF-TssG baseplate subcomplex revealed by cryo-electron microscopy - full map sharpened | | 分子名称: | Putative type VI secretion protein, Unassigned protein | | 著者 | Park, Y.J, Lacourse, K.D, Cambillau, C, Seattle Structural Genomics Center for Infectious Disease (SSGCID), DiMaio, F, Mougous, J.D, Veesler, D. | | 登録日 | 2018-11-14 | | 公開日 | 2018-12-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure of the type VI secretion system TssK-TssF-TssG baseplate subcomplex revealed by cryo-electron microscopy.
Nat Commun, 9, 2018
|
|
6BQM
 
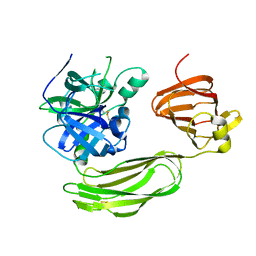 | | Secreted serine protease VesC from Vibrio cholerae | | 分子名称: | serine protease VesC | | 著者 | Park, Y.J, Korotkov, K.V, Delarosa, J.R, Turley, S, DiMaio, F, Hol, W.G.J. | | 登録日 | 2017-11-28 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Suppressor Mutations in Type II Secretion Mutants of Vibrio cholerae: Inactivation of the VesC Protease.
Msphere, 5, 2020
|
|
6WPT
 
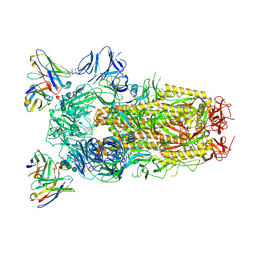 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment (open state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | 著者 | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2020-04-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
